Gene Page: GPT
Summary ?
| GeneID | 2875 |
| Symbol | GPT |
| Synonyms | AAT1|ALT1|GPT1 |
| Description | glutamic-pyruvate transaminase (alanine aminotransferase) |
| Reference | MIM:138200|HGNC:HGNC:4552|Ensembl:ENSG00000167701|HPRD:00689|Vega:OTTHUMG00000165176 |
| Gene type | protein-coding |
| Map location | 8q24.3 |
| Pascal p-value | 0.135 |
| Fetal beta | -1.228 |
| DMG | 1 (# studies) |
| eGene | Cerebellum |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17869161 | 8 | 145729373 | GPT | 1.662E-4 | -0.337 | 0.033 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
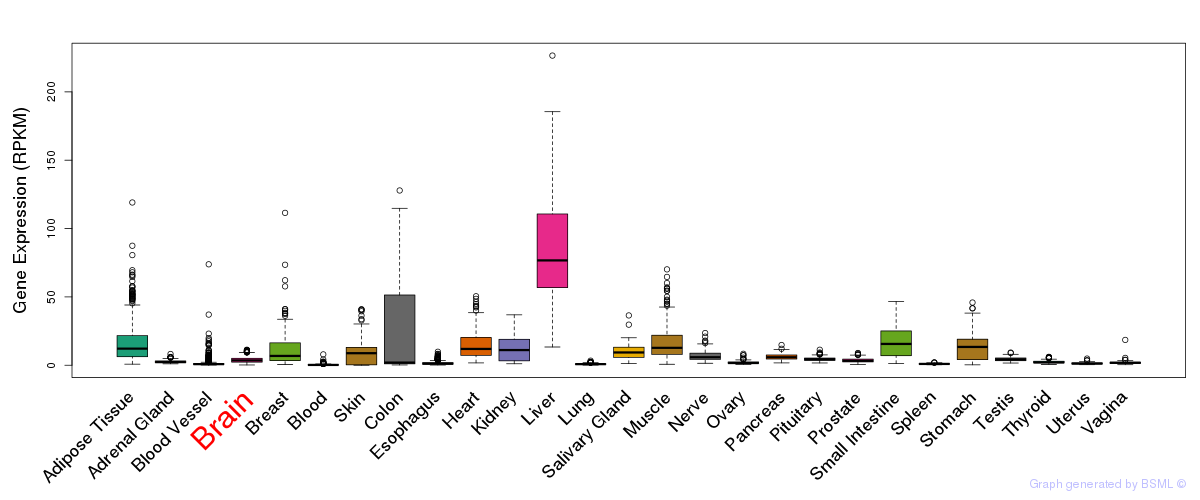
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PRDM2 | 0.85 | 0.88 |
| RAPGEF2 | 0.85 | 0.89 |
| SBNO1 | 0.84 | 0.87 |
| TEX2 | 0.84 | 0.87 |
| PRKCE | 0.83 | 0.87 |
| SDAD1 | 0.83 | 0.87 |
| ADRBK2 | 0.83 | 0.88 |
| CAMK1D | 0.83 | 0.85 |
| UBP1 | 0.82 | 0.85 |
| GRIA2 | 0.82 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RHOC | -0.63 | -0.69 |
| AF347015.21 | -0.62 | -0.65 |
| ENHO | -0.61 | -0.71 |
| S100A16 | -0.61 | -0.65 |
| HIGD1B | -0.61 | -0.64 |
| AF347015.31 | -0.60 | -0.63 |
| RAB34 | -0.60 | -0.65 |
| MT-CO2 | -0.60 | -0.62 |
| C1orf54 | -0.60 | -0.71 |
| PLA2G5 | -0.60 | -0.65 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004021 | alanine transaminase activity | EXP | glutamate (GO term level: 6) | 9119391 |
| GO:0004021 | alanine transaminase activity | NAS | glutamate (GO term level: 6) | 1931970 |
| GO:0016847 | 1-aminocyclopropane-1-carboxylate synthase activity | IEA | - | |
| GO:0030170 | pyridoxal phosphate binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006094 | gluconeogenesis | NAS | - | |
| GO:0009058 | biosynthetic process | IEA | - | |
| GO:0006807 | nitrogen compound metabolic process | NAS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 9119391 | |
| GO:0005737 | cytoplasm | NAS | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ALANINE ASPARTATE AND GLUTAMATE METABOLISM | 32 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SARS PATHWAY | 10 | 5 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | 17 | 14 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q23 Q24 AMPLICON | 157 | 87 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| SU LIVER | 55 | 32 | All SZGR 2.0 genes in this pathway |
| SCHLINGEMANN SKIN CARCINOGENESIS TPA DN | 29 | 15 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION DN | 179 | 97 | All SZGR 2.0 genes in this pathway |
| COULOUARN TEMPORAL TGFB1 SIGNATURE DN | 138 | 99 | All SZGR 2.0 genes in this pathway |
| WOO LIVER CANCER RECURRENCE DN | 80 | 54 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA FGF2 TARGETS | 29 | 17 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| MOOTHA GLUCONEOGENESIS | 32 | 23 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |