Gene Page: OLA1
Summary ?
| GeneID | 29789 |
| Symbol | OLA1 |
| Synonyms | DOC45|GBP45|GTBP9|GTPBP9|PTD004 |
| Description | Obg-like ATPase 1 |
| Reference | MIM:611175|HGNC:HGNC:28833|Ensembl:ENSG00000138430|HPRD:15194|Vega:OTTHUMG00000132335 |
| Gene type | protein-coding |
| Map location | 2q31.1 |
| Pascal p-value | 0.002 |
| Sherlock p-value | 0.796 |
| eGene | Myers' cis & trans Meta |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17317002 | chr2 | 174954300 | OLA1 | 29789 | 0.04 | cis | ||
| rs10930638 | chr2 | 174974595 | OLA1 | 29789 | 1.395E-8 | cis | ||
| rs17239377 | chr2 | 174980338 | OLA1 | 29789 | 4.689E-7 | cis | ||
| rs6433464 | chr2 | 175008771 | OLA1 | 29789 | 1.757E-7 | cis | ||
| rs11674895 | chr2 | 175013962 | OLA1 | 29789 | 3.965E-7 | cis | ||
| rs10195413 | chr2 | 175033173 | OLA1 | 29789 | 1.699E-7 | cis | ||
| rs4972643 | chr2 | 175059700 | OLA1 | 29789 | 1.573E-7 | cis | ||
| rs10930654 | chr2 | 175063512 | OLA1 | 29789 | 5.343E-7 | cis | ||
| rs4144329 | chr2 | 175070877 | OLA1 | 29789 | 5.343E-7 | cis | ||
| rs10930656 | chr2 | 175117879 | OLA1 | 29789 | 0.02 | cis | ||
| rs12470835 | chr2 | 175120198 | OLA1 | 29789 | 5.48E-4 | cis | ||
| rs10930638 | chr2 | 174974595 | OLA1 | 29789 | 3.294E-6 | trans | ||
| rs17239377 | chr2 | 174980338 | OLA1 | 29789 | 9.325E-5 | trans | ||
| rs6433464 | chr2 | 175008771 | OLA1 | 29789 | 3.809E-5 | trans | ||
| rs11674895 | chr2 | 175013962 | OLA1 | 29789 | 8.001E-5 | trans | ||
| rs10195413 | chr2 | 175033173 | OLA1 | 29789 | 3.689E-5 | trans | ||
| rs4972643 | chr2 | 175059700 | OLA1 | 29789 | 3.41E-5 | trans | ||
| rs10930654 | chr2 | 175063512 | OLA1 | 29789 | 1.051E-4 | trans | ||
| rs4144329 | chr2 | 175070877 | OLA1 | 29789 | 1.051E-4 | trans | ||
| rs12470835 | chr2 | 175120198 | OLA1 | 29789 | 0.04 | trans |
Section II. Transcriptome annotation
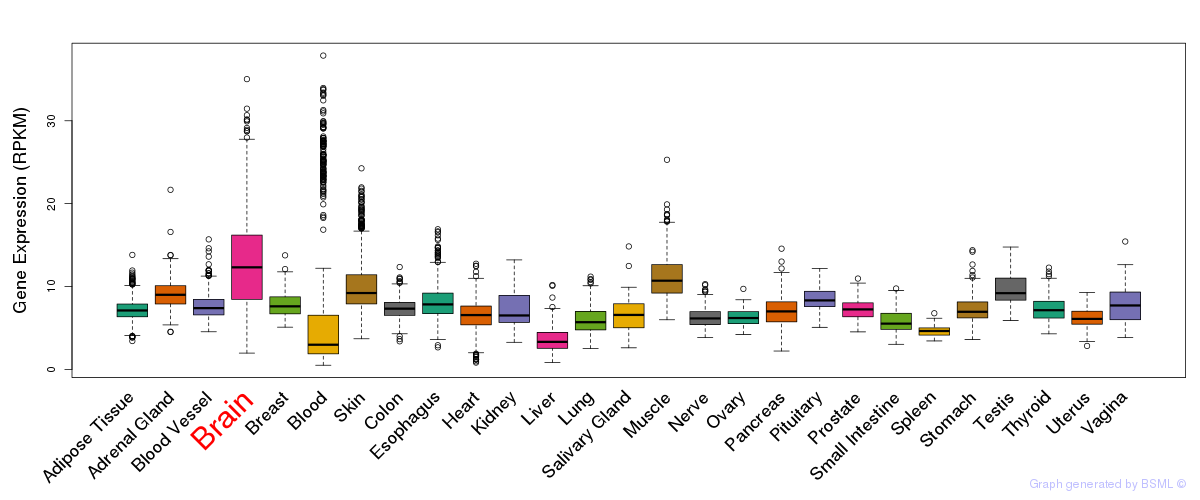
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 DN | 116 | 71 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT UP | 197 | 110 | All SZGR 2.0 genes in this pathway |
| KENNY CTNNB1 TARGETS DN | 52 | 34 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO UP | 205 | 126 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| GRADE METASTASIS DN | 45 | 31 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| WINNEPENNINCKX MELANOMA METASTASIS UP | 162 | 86 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM SENSITIVE GENES | 90 | 54 | All SZGR 2.0 genes in this pathway |
| BILANGES RAPAMYCIN SENSITIVE VIA TSC1 AND TSC2 | 73 | 37 | All SZGR 2.0 genes in this pathway |
| VANOEVELEN MYOGENESIS SIN3A TARGETS | 220 | 133 | All SZGR 2.0 genes in this pathway |