Gene Page: HTT
Summary ?
| GeneID | 3064 |
| Symbol | HTT |
| Synonyms | HD|IT15 |
| Description | huntingtin |
| Reference | MIM:613004|HGNC:HGNC:4851|Ensembl:ENSG00000197386|HPRD:00883|Vega:OTTHUMG00000159916 |
| Gene type | protein-coding |
| Map location | 4p16.3 |
| Pascal p-value | 0.59 |
| Sherlock p-value | 0.915 |
| Fetal beta | 0.017 |
| eGene | Frontal Cortex BA9 Myers' cis & trans |
| Support | CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 6.0206 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HTT | chr4 | 3215762 | G | A | NM_002111 | . | silent | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10497672 | 0 | HTT | 3064 | 0.02 | trans |
Section II. Transcriptome annotation
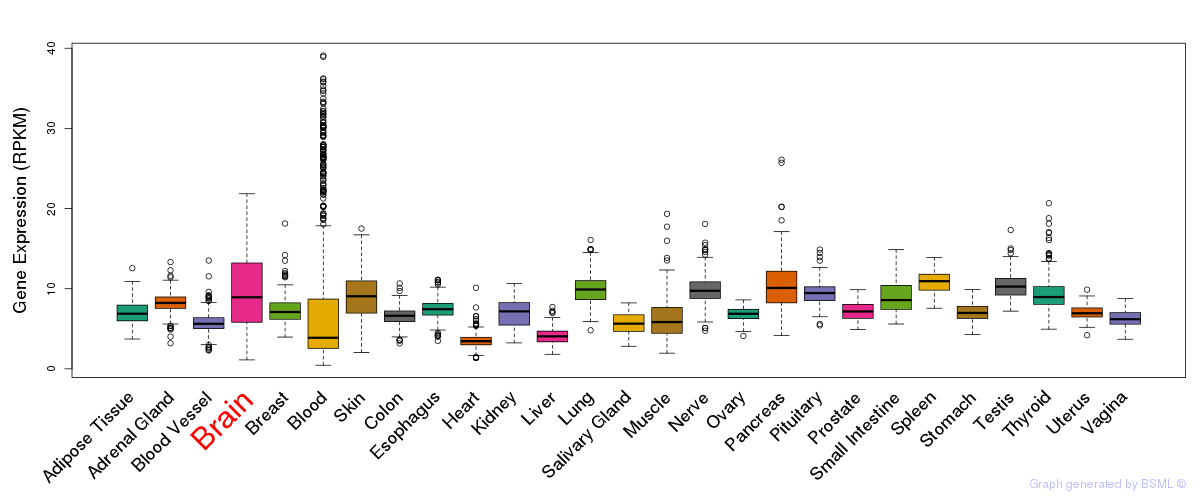
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DIO2 | 0.76 | 0.69 |
| SLC1A3 | 0.73 | 0.69 |
| GLUL | 0.70 | 0.56 |
| SLCO1C1 | 0.68 | 0.60 |
| C4orf19 | 0.67 | 0.55 |
| MERTK | 0.65 | 0.56 |
| ACSS3 | 0.64 | 0.57 |
| PPP1R3C | 0.64 | 0.59 |
| HEPH | 0.63 | 0.50 |
| LYN | 0.63 | 0.63 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TSPAN2 | -0.33 | -0.34 |
| SNTG2 | -0.31 | -0.37 |
| RAB33A | -0.30 | -0.30 |
| AC110814.1 | -0.30 | -0.28 |
| SDC1 | -0.30 | -0.29 |
| EFNA2 | -0.30 | -0.34 |
| MED19 | -0.30 | -0.31 |
| DPF1 | -0.30 | -0.22 |
| RNF19B | -0.29 | -0.24 |
| FAM110A | -0.29 | -0.28 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003714 | transcription corepressor activity | TAS | 10823891 | |
| GO:0005515 | protein binding | IPI | 7477378 | |
| GO:0005215 | transporter activity | TAS | 7568002 | |
| GO:0008017 | microtubule binding | TAS | 7568002 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006917 | induction of apoptosis | TAS | 9778247 | |
| GO:0009887 | organ morphogenesis | TAS | 7774020 | |
| GO:0009405 | pathogenesis | TAS | 10353249 | |
| GO:0007610 | behavior | TAS | 7774020 | |
| GO:0006915 | apoptosis | TAS | 8696339 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005794 | Golgi apparatus | IDA | 15837803 | |
| GO:0005625 | soluble fraction | TAS | 7568002 | |
| GO:0005634 | nucleus | TAS | 9778247 | |
| GO:0005737 | cytoplasm | TAS | 7568002 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | Huntingtin is serine-phosphorylated by Akt. | BIND | 12062094 |
| AP2A2 | ADTAB | CLAPA2 | HIP9 | HYPJ | adaptor-related protein complex 2, alpha 2 subunit | Two-hybrid | BioGRID | 9700202 |
| CASP1 | ICE | IL1BC | P45 | caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) | Huntingtin interacts with caspase 1. This interaction was modeled on a demonstrated interaction between human huntingtin and caspase 1 from an unspecified species. | BIND | 9535906 |
| CASP1 | ICE | IL1BC | P45 | caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) | - | HPRD | 9535906 |
| CASP2 | CASP-2 | ICH-1L | ICH-1L/1S | ICH1 | NEDD2 | caspase 2, apoptosis-related cysteine peptidase | Htt interacts with caspase 2. | BIND | 14713958 |
| CASP3 | CPP32 | CPP32B | SCA-1 | caspase 3, apoptosis-related cysteine peptidase | Huntingtin interacts with caspase 3. This interaction was modeled on a demonstrated interaction between human Huntingtin and caspase 3 from an unspecified species. | BIND | 9535906 |
| CASP6 | MCH2 | caspase 6, apoptosis-related cysteine peptidase | Htt interacts with caspase 6. | BIND | 14713958 |
| CASP7 | CMH-1 | ICE-LAP3 | MCH3 | caspase 7, apoptosis-related cysteine peptidase | Htt interacts with caspase 7. | BIND | 14713958 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | Htt interacts with caspase 8. | BIND | 14713958 |
| CBS | HIP4 | cystathionine-beta-synthase | - | HPRD | 10434301 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | CBP interacts with htt. This interaction was modeled on a demonstrated interaction between CBP and htt from unspecified species. | BIND | 10823891 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | - | HPRD,BioGRID | 11264541 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | Htt interacts with CBP. | BIND | 15225551 |
| CTBP1 | BARS | MGC104684 | C-terminal binding protein 1 | Huntingtin interacts with CTBP. | BIND | 11739372 |
| CTBP1 | BARS | MGC104684 | C-terminal binding protein 1 | - | HPRD,BioGRID | 11739372 |
| CXorf27 | 1700054O13Rik | HIP17 | HYPM | chromosome X open reading frame 27 | Two-hybrid | BioGRID | 9700202 |
| DCTN1 | DAP-150 | DP-150 | HMN7B | P135 | dynactin 1 (p150, glued homolog, Drosophila) | - | HPRD | 9454836 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | Huntingtin interacts with PSD-95. | BIND | 11319238 |
| DNAJB2 | HSJ1 | HSPF3 | DnaJ (Hsp40) homolog, subfamily B, member 2 | HSJ1a interacts with Htt-Q103. | BIND | 15936278 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD,BioGRID | 9079622 |
| EIF2AK2 | EIF2AK1 | MGC126524 | PKR | PRKR | eukaryotic translation initiation factor 2-alpha kinase 2 | PKR interacts with huntingtin mRNA. | BIND | 11468270 |
| F8A1 | DXS522E | F8A | HAP40 | coagulation factor VIII-associated (intronic transcript) 1 | - | HPRD,BioGRID | 11035034 |
| FICD | HIP13 | HYPE | MGC5623 | UNQ3041 | FIC domain containing | - | HPRD,BioGRID | 9700202 |
| GAPDH | G3PD | GAPD | MGC88685 | glyceraldehyde-3-phosphate dehydrogenase | - | HPRD,BioGRID | 8612237 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD,BioGRID | 9079622 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | HDP interacts with Grb2. | BIND | 9079622 |
| HAP1 | HAP2 | HIP5 | HLP | hHLP1 | huntingtin-associated protein 1 | Affinity Capture-Western in vitro in vivo Reconstituted Complex Two-hybrid | BioGRID | 7477378 |9454836 |9599014 |9668110 |
| HAP1 | HAP2 | HIP5 | HLP | hHLP1 | huntingtin-associated protein 1 | Htt interacts with HAP1. This interaction was modeled on a demonstrated interaction between human Htt and Rat HAP1. | BIND | 15654337 |
| HAP1 | HAP2 | HIP5 | HLP | hHLP1 | huntingtin-associated protein 1 | Huntingtin interacts with HAP1. | BIND | 9668110 |
| HAP1 | HAP2 | HIP5 | HLP | hHLP1 | huntingtin-associated protein 1 | Htt interacts with Hap1. | BIND | 7477378 |
| HAP1 | HAP2 | HIP5 | HLP | hHLP1 | huntingtin-associated protein 1 | - | HPRD | 7477378 |9599014 |9668110 |10974549 |
| HIP1 | ILWEQ | MGC126506 | huntingtin interacting protein 1 | - | HPRD,BioGRID | 9147654 |11063258 |
| HIP1 | ILWEQ | MGC126506 | huntingtin interacting protein 1 | HIP1 interacts with HD. | BIND | 11063258 |
| HIP1R | FLJ14000 | FLJ27022 | HIP12 | HIP3 | ILWEQ | KIAA0655 | MGC47513 | huntingtin interacting protein 1 related | - | HPRD | 11063258 |
| HSPA4 | APG-2 | HS24/P52 | MGC131852 | RY | hsp70 | hsp70RY | heat shock 70kDa protein 4 | Htt interacts with HSP70. | BIND | 15654337 |
| HTT | HD | IT15 | huntingtin | Htt interacts with itself to form oligomers. | BIND | 15225551 |
| HYPK | HSPC136 | Huntingtin interacting protein K | - | HPRD,BioGRID | 9700202 |
| IFT57 | ESRRBL1 | FLJ10147 | HIPPI | MHS4R2 | intraflagellar transport 57 homolog (Chlamydomonas) | - | HPRD | 11788820 |
| MAGEA3 | HIP8 | HYPD | MAGE3 | MAGEA6 | MGC14613 | melanoma antigen family A, 3 | Two-hybrid | BioGRID | 9700202 |
| MAP3K10 | MLK2 | MST | mitogen-activated protein kinase kinase kinase 10 | MLK2 SH3 domain interacts with Huntingtin | BIND | 10801775 |
| MAP3K10 | MLK2 | MST | mitogen-activated protein kinase kinase kinase 10 | - | HPRD,BioGRID | 10801775 |
| MED31 | 3110004H13Rik | CGI-125 | FLJ27436 | FLJ36714 | Soh1 | mediator complex subunit 31 | - | HPRD | 15383276 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | HD interacts with N-CoR. | BIND | 10441327 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | Two-hybrid | BioGRID | 10441327 |
| NEUROD1 | BETA2 | BHF-1 | NEUROD | bHLHa3 | neurogenic differentiation 1 | Affinity Capture-Western | BioGRID | 12881483 |
| OPTN | FIP2 | GLC1E | HIP7 | HYPL | NRP | TFIIIA-INTP | optineurin | FIP-2 carboxy-terminus interacts with Huntingtin in perinuclear vesicular structures. | BIND | 11137014 |
| OPTN | FIP2 | GLC1E | HIP7 | HYPL | NRP | TFIIIA-INTP | optineurin | - | HPRD,BioGRID | 11137014 |
| PACSIN1 | KIAA1379 | SDPI | protein kinase C and casein kinase substrate in neurons 1 | - | HPRD,BioGRID | 12354780 |
| PACSIN1 | KIAA1379 | SDPI | protein kinase C and casein kinase substrate in neurons 1 | huntingtin interacts with PACSIN1. | BIND | 12354780 |
| PRPF40A | FBP-11 | FBP11 | FLAF1 | FLJ20585 | FNBP3 | HIP10 | HYPA | NY-REN-6 | PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) | - | HPRD,BioGRID | 9700202 |
| PRPF40A | FBP-11 | FBP11 | FLAF1 | FLJ20585 | FNBP3 | HIP10 | HYPA | NY-REN-6 | PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) | Huntingtin interacts with HYPA | BIND | 9700202 |
| PRPF40B | HYPC | PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) | Huntingtin interacts with HYPC | BIND | 9700202 |
| PRPF40B | HYPC | PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) | - | HPRD,BioGRID | 9700202 |
| RASA1 | CM-AVM | CMAVM | DKFZp434N071 | GAP | PKWS | RASA | RASGAP | p120GAP | p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 | HDP interacts with RasGAP. | BIND | 9079622 |
| RASA1 | CM-AVM | CMAVM | DKFZp434N071 | GAP | PKWS | RASA | RASGAP | p120GAP | p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 | - | HPRD,BioGRID | 9079622 |
| SETD2 | FLJ16420 | FLJ22472 | FLJ23184 | FLJ45883 | FLJ46217 | HIF-1 | HSPC069 | HYPB | KIAA1732 | KMT3A | SET2 | p231HBP | SET domain containing 2 | - | HPRD,BioGRID | 9700202 |
| SETD2 | FLJ16420 | FLJ22472 | FLJ23184 | FLJ45883 | FLJ46217 | HIF-1 | HSPC069 | HYPB | KIAA1732 | KMT3A | SET2 | p231HBP | SET domain containing 2 | Huntingtin interacts with HYPB | BIND | 9700202 |
| SH3GL3 | CNSA3 | EEN-2B-L3 | EEN-B2 | HsT19371 | SH3D2C | SH3P13 | SH3-domain GRB2-like 3 | HD interacts with SH3GL3. | BIND | 9809064 |
| SH3GL3 | CNSA3 | EEN-2B-L3 | EEN-B2 | HsT19371 | SH3D2C | SH3P13 | SH3-domain GRB2-like 3 | - | HPRD,BioGRID | 9809064 |
| SH3GLB1 | Bif-1 | CGI-61 | KIAA0491 | dJ612B15.2 | SH3-domain GRB2-like endophilin B1 | - | HPRD,BioGRID | 12456676 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | mSin3A interacts with htt. This interaction was modeled on a demonstrated interaction between mSin3A and htt from unspecified species. | BIND | 10823891 |
| SP1 | - | Sp1 transcription factor | - | HPRD,BioGRID | 11839795 |
| SP1 | - | Sp1 transcription factor | Huntingtin interacts with Sp1. This interaction was modelled on a demonstrated interaction between Huntingtin from human and Sp1 from rat. | BIND | 11839795 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | HD is covalently attached to SUMO-1 via lysine residues. | BIND | 15064418 |
| TAF4 | FLJ41943 | TAF2C | TAF2C1 | TAF4A | TAFII130 | TAFII135 | TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa | TAFII130 interacts with htt. | BIND | 11988536 |
| TAF4 | FLJ41943 | TAF2C | TAF2C1 | TAF4A | TAFII130 | TAFII135 | TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa | - | HPRD | 11988536 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | Htt interacts with TBP. | BIND | 15225551 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | - | HPRD | 10410676 |
| TCERG1 | CA150 | MGC133200 | TAF2S | transcription elongation regulator 1 | Htt interacts with CA150. | BIND | 11172033 |
| TCERG1 | CA150 | MGC133200 | TAF2S | transcription elongation regulator 1 | - | HPRD,BioGRID | 11172033 |
| TGM2 | G-ALPHA-h | GNAH | TG2 | TGC | transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) | Htt interacts with tTG. | BIND | 11442349 |
| TGM2 | G-ALPHA-h | GNAH | TG2 | TGC | transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) | - | HPRD | 11442349 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p53 interacts with htt. This interaction was modeled on a demonstrated interaction between p53 from human and htt from an unspecified species. | BIND | 10823891 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Htt interacts with TP53(p53). | BIND | 15996546 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 10823891 |
| TPH1 | MGC119994 | TPH | TPRH | tryptophan hydroxylase 1 | - | HPRD | 12354289 |
| TPR | - | translocated promoter region (to activated MET oncogene) | Htt interacts with Tpr. | BIND | 15654337 |
| TRIP10 | CIP4 | HSTP | STOT | STP | thyroid hormone receptor interactor 10 | htt interacts with CIP4. | BIND | 12604778 |
| TRIP10 | CIP4 | HSTP | STOT | STP | thyroid hormone receptor interactor 10 | - | HPRD,BioGRID | 12604778 |
| UBE2K | DKFZp564C1216 | DKFZp686J24237 | E2-25K | HIP2 | HYPG | LIG | ubiquitin-conjugating enzyme E2K (UBC1 homolog, yeast) | - | HPRD,BioGRID | 8702625 |
| UBE2K | DKFZp564C1216 | DKFZp686J24237 | E2-25K | HIP2 | HYPG | LIG | ubiquitin-conjugating enzyme E2K (UBC1 homolog, yeast) | HD interacts with hE2-25K. | BIND | 8702625 |
| ZDHHC17 | HIP14 | HIP3 | HSPC294 | HYPH | KIAA0946 | zinc finger, DHHC-type containing 17 | HIP14 catalyzes palmitoylation of htt. This interaction was modeled on a demonstrated interaction between human HIP14 and htt from an unspecified species. | BIND | 15603740 |
| ZDHHC17 | HIP14 | HIP3 | HSPC294 | HYPH | KIAA0946 | zinc finger, DHHC-type containing 17 | - | HPRD,BioGRID | 9700202 |12393793 |
| ZDHHC17 | HIP14 | HIP3 | HSPC294 | HYPH | KIAA0946 | zinc finger, DHHC-type containing 17 | htt interacts with HIP14. | BIND | 12393793 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS BLUE DN | 61 | 35 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE UP | 134 | 93 | All SZGR 2.0 genes in this pathway |
| SCHEIDEREIT IKK INTERACTING PROTEINS | 58 | 45 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR ANAPLASTIC UP | 19 | 12 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| MANTOVANI VIRAL GPCR SIGNALING DN | 49 | 37 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND TBH | 60 | 42 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 UP | 167 | 99 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE UP | 163 | 102 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS VS STROMAL STIMULATION DN | 99 | 65 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-137 | 3096 | 3102 | 1A | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-216 | 582 | 588 | 1A | hsa-miR-216 | UAAUCUCAGCUGGCAACUGUG |
| miR-411 | 613 | 619 | 1A | hsa-miR-411 | AACACGGUCCACUAACCCUCAGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.