Gene Page: HUNK
Summary ?
| GeneID | 30811 |
| Symbol | HUNK |
| Synonyms | - |
| Description | hormonally up-regulated Neu-associated kinase |
| Reference | MIM:606532|HGNC:HGNC:13326|Ensembl:ENSG00000142149|HPRD:09410|Vega:OTTHUMG00000085019 |
| Gene type | protein-coding |
| Map location | 21q22.1 |
| Pascal p-value | 0.029 |
| Fetal beta | 1.32 |
| DMG | 1 (# studies) |
| eGene | Meta |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg11173579 | 21 | 33247885 | HUNK | 4.59E-5 | 0.686 | 0.021 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
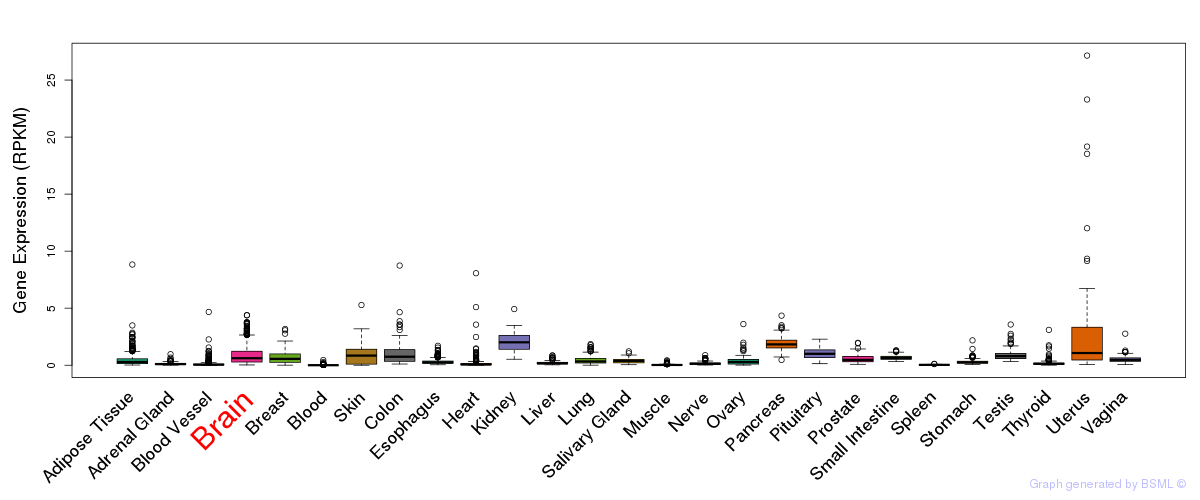
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CLDN5 | 0.60 | 0.52 |
| SH3TC1 | 0.58 | 0.48 |
| ICAM2 | 0.57 | 0.50 |
| C10orf10 | 0.56 | 0.35 |
| HRCT1 | 0.53 | 0.20 |
| IL32 | 0.51 | 0.34 |
| NFKB2 | 0.51 | 0.45 |
| IGFBP1 | 0.50 | 0.25 |
| EDN1 | 0.49 | 0.26 |
| TMEM204 | 0.48 | 0.36 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZNF540 | -0.41 | -0.46 |
| CDH10 | -0.40 | -0.47 |
| RNF103 | -0.40 | -0.44 |
| MYLIP | -0.40 | -0.50 |
| Z68873.1 | -0.39 | -0.41 |
| AC141586.1 | -0.39 | -0.40 |
| ZFAND5 | -0.38 | -0.46 |
| TAOK3 | -0.38 | -0.37 |
| AFTPH | -0.37 | -0.41 |
| CCDC149 | -0.37 | -0.41 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 AND TP63 TARGETS | 205 | 145 | All SZGR 2.0 genes in this pathway |
| LIAO HAVE SOX4 BINDING SITES | 40 | 26 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS UP | 126 | 84 | All SZGR 2.0 genes in this pathway |
| VERNELL RETINOBLASTOMA PATHWAY UP | 70 | 47 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS | 212 | 121 | All SZGR 2.0 genes in this pathway |
| CERIBELLI PROMOTERS INACTIVE AND BOUND BY NFY | 44 | 20 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |