Gene Page: HSD3B1
Summary ?
| GeneID | 3283 |
| Symbol | HSD3B1 |
| Synonyms | 3BETAHSD|HSD3B|HSDB3|HSDB3A|I|SDR11E1 |
| Description | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| Reference | MIM:109715|HGNC:HGNC:5217|Ensembl:ENSG00000203857|HPRD:04920|Vega:OTTHUMG00000012525 |
| Gene type | protein-coding |
| Map location | 1p13.1 |
| Pascal p-value | 0.314 |
| Fetal beta | -0.034 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
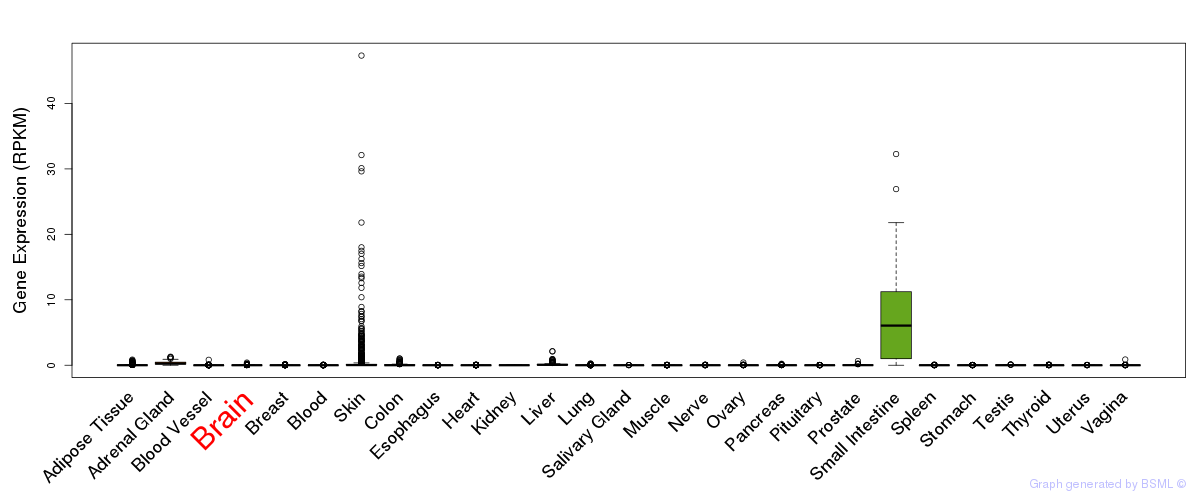
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HSPA1B | 0.98 | 0.75 |
| HSPA6 | 0.94 | 0.56 |
| DNAJB1 | 0.92 | 0.60 |
| G0S2 | 0.85 | 0.39 |
| BAG3 | 0.82 | 0.46 |
| AL031847.1 | 0.67 | 0.04 |
| ACRC | 0.65 | 0.04 |
| HSPB1 | 0.64 | 0.46 |
| SERPINH1 | 0.56 | 0.51 |
| GPR84 | 0.56 | 0.29 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MFSD5 | -0.23 | -0.03 |
| IFNAR2 | -0.21 | -0.07 |
| DOLK | -0.20 | 0.03 |
| MAN1A2 | -0.20 | -0.16 |
| ZNF706 | -0.19 | -0.11 |
| CDC42EP2 | -0.19 | -0.03 |
| C19orf39 | -0.19 | -0.12 |
| TMEM86A | -0.19 | -0.04 |
| MYLK2 | -0.19 | -0.14 |
| TRAM1L1 | -0.19 | -0.08 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity | IEA | - | |
| GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity | ISS | 1944309 | |
| GO:0005488 | binding | IEA | - | |
| GO:0004769 | steroid delta-isomerase activity | ISS | 1944309 | |
| GO:0016853 | isomerase activity | IEA | - | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006703 | estrogen biosynthetic process | TAS | 1944309 | |
| GO:0006702 | androgen biosynthetic process | TAS | 1944309 | |
| GO:0006700 | C21-steroid hormone biosynthetic process | IEA | - | |
| GO:0006694 | steroid biosynthetic process | IEA | - | |
| GO:0055114 | oxidation reduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005792 | microsome | IDA | 8274411 | |
| GO:0005739 | mitochondrion | IEA | - | |
| GO:0005743 | mitochondrial inner membrane | IDA | 8274411 | |
| GO:0005758 | mitochondrial intermembrane space | IDA | 8274411 | |
| GO:0005783 | endoplasmic reticulum | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0030868 | smooth endoplasmic reticulum membrane | ISS | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG STEROID HORMONE BIOSYNTHESIS | 55 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | 35 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME ANDROGEN BIOSYNTHESIS | 10 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME STEROID HORMONES | 29 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO UP | 205 | 126 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS UP | 88 | 47 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS UP | 102 | 64 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS UP | 91 | 59 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN UP | 90 | 58 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |
| ROESSLER LIVER CANCER METASTASIS UP | 107 | 72 | All SZGR 2.0 genes in this pathway |