Gene Page: HSP90AA1
Summary ?
| GeneID | 3320 |
| Symbol | HSP90AA1 |
| Synonyms | EL52|HEL-S-65p|HSP86|HSP89A|HSP90A|HSP90N|HSPC1|HSPCA|HSPCAL1|HSPCAL4|HSPN|Hsp89|Hsp90|LAP-2|LAP2 |
| Description | heat shock protein 90kDa alpha family class A member 1 |
| Reference | MIM:140571|HGNC:HGNC:5253|Ensembl:ENSG00000080824|HPRD:00777|Vega:OTTHUMG00000171752 |
| Gene type | protein-coding |
| Map location | 14q32.33 |
| Pascal p-value | 0.299 |
| TADA p-value | 0.002 |
| Fetal beta | -0.198 |
| DMG | 2 (# studies) |
| Support | RNA AND PROTEIN SYNTHESIS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 4 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 4 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.4681 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HSP90AA1 | chr14 | 102550757 | C | CCT | NM_001017963 NM_005348 | . . | frameshift frameshift | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg13797425 | 14 | 102554977 | HSP90AA1 | 1.5E-7 | 0.741 | 0.004 | DMG:Wockner_2014 |
| cg14893857 | 14 | 102554969 | HSP90AA1 | 3.67E-5 | 0.47 | 0.02 | DMG:Wockner_2014 |
| cg23289024 | 14 | 102554846 | HSP90AA1 | 5.143E-4 | 0.471 | 0.047 | DMG:Wockner_2014 |
| cg04281268 | 14 | 102606293 | HSP90AA1 | -0.02 | 0.96 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
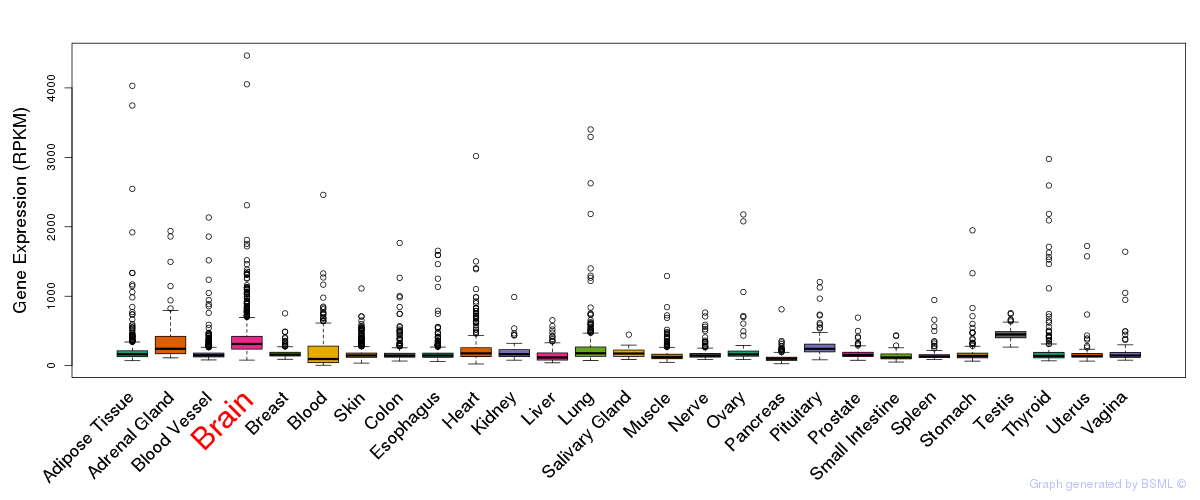
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MON1B | 0.90 | 0.93 |
| UPF1 | 0.90 | 0.91 |
| CLEC16A | 0.90 | 0.91 |
| ZFYVE20 | 0.90 | 0.90 |
| BICD2 | 0.89 | 0.91 |
| POM121 | 0.89 | 0.89 |
| DGCR2 | 0.89 | 0.90 |
| PCNXL3 | 0.89 | 0.88 |
| CLPB | 0.89 | 0.89 |
| TULP4 | 0.89 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.67 | -0.75 |
| C1orf54 | -0.64 | -0.74 |
| AF347015.31 | -0.64 | -0.66 |
| GNG11 | -0.63 | -0.68 |
| MT-CO2 | -0.61 | -0.65 |
| SYCP3 | -0.60 | -0.69 |
| HIGD1B | -0.60 | -0.63 |
| VAMP5 | -0.59 | -0.61 |
| AF347015.27 | -0.59 | -0.63 |
| IFI27 | -0.58 | -0.58 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | TAS | 11470816 | |
| GO:0003674 | molecular_function | ND | - | |
| GO:0030235 | nitric-oxide synthase regulator activity | ISS | - | |
| GO:0005524 | ATP binding | TAS | 11470816 | |
| GO:0051082 | unfolded protein binding | IEA | - | |
| GO:0042803 | protein homodimerization activity | TAS | 11470816 | |
| GO:0030911 | TPR domain binding | IDA | 9660753 | |
| GO:0030911 | TPR domain binding | TAS | 12526792 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006839 | mitochondrial transport | TAS | glutamate (GO term level: 6) | 12526792 |
| GO:0006457 | protein folding | IEA | - | |
| GO:0006986 | response to unfolded protein | NAS | 2527334 | |
| GO:0007165 | signal transduction | NAS | 11470816 | |
| GO:0008150 | biological_process | ND | - | |
| GO:0042026 | protein refolding | TAS | 9660753 | |
| GO:0045429 | positive regulation of nitric oxide biosynthetic process | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 9580552 |10781589 | |
| GO:0005829 | cytosol | NAS | 12526792 | |
| GO:0005575 | cellular_component | ND | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0042470 | melanosome | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AHR | - | aryl hydrocarbon receptor | - | HPRD,BioGRID | 7961671 |9083006 |
| AHSA1 | AHA1 | C14orf3 | p38 | AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast) | in vitro in vivo Two-hybrid | BioGRID | 12504007 |
| AHSA1 | AHA1 | C14orf3 | p38 | AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast) | - | HPRD | 12604615 |
| AHSA2 | DKFZp564C236 | FLJ34679 | FLJ41715 | Hch1 | AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast) | - | HPRD,BioGRID | 12504007 |14739935 |
| AIP | ARA9 | FKBP16 | FKBP37 | SMTPHN | XAP2 | aryl hydrocarbon receptor interacting protein | - | HPRD | 9111057 |9837941 |
| AIP | ARA9 | FKBP16 | FKBP37 | SMTPHN | XAP2 | aryl hydrocarbon receptor interacting protein | Affinity Capture-Western | BioGRID | 11013261 |
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | - | HPRD,BioGRID | 10995457 |
| AKT2 | PKBB | PKBBETA | PRKBB | RAC-BETA | v-akt murine thymoma viral oncogene homolog 2 | - | HPRD | 11988487 |
| ALK | CD246 | Ki-1 | TFG/ALK | anaplastic lymphoma receptor tyrosine kinase | - | HPRD | 11888936 |
| APAF1 | CED4 | DKFZp781B1145 | apoptotic peptidase activating factor 1 | Affinity Capture-Western | BioGRID | 10934467 |
| APOB | FLDB | apolipoprotein B (including Ag(x) antigen) | - | HPRD | 11333259 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD,BioGRID | 1525041 |
| ARNT | HIF-1beta | HIF1B | HIF1BETA | TANGO | bHLHe2 | aryl hydrocarbon receptor nuclear translocator | Affinity Capture-Western | BioGRID | 11013261 |
| ARNTL | BMAL1 | BMAL1c | JAP3 | MGC47515 | MOP3 | PASD3 | TIC | bHLHe5 | aryl hydrocarbon receptor nuclear translocator-like | - | HPRD,BioGRID | 9079689 |
| ASGR1 | ASGPR | CLEC4H1 | Hs.12056 | asialoglycoprotein receptor 1 | - | HPRD | 12167617 |
| BAG4 | BAG-4 | SODD | BCL2-associated athanogene 4 | Affinity Capture-Western | BioGRID | 14559896 |
| BIRC5 | API4 | EPR-1 | baculoviral IAP repeat-containing 5 | Survivin interacts with Hsp90. | BIND | 15894266 |
| CALM1 | CALML2 | CAMI | DD132 | PHKD | calmodulin 1 (phosphorylase kinase, delta) | - | HPRD | 8486648 |
| CDC37 | P50CDC37 | cell division cycle 37 homolog (S. cerevisiae) | Co-crystal Structure in vitro | BioGRID | 9685350 |14718169 |
| CDC37 | P50CDC37 | cell division cycle 37 homolog (S. cerevisiae) | - | HPRD | 9685350|14718169 |
| CHTF18 | C16orf41 | C321D2.2 | C321D2.3 | C321D2.4 | CHL12 | Ctf18 | RUVBL | CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae) | Affinity Capture-MS | BioGRID | 12930902 |
| CHUK | IKBKA | IKK-alpha | IKK1 | IKKA | NFKBIKA | TCF16 | conserved helix-loop-helix ubiquitous kinase | - | HPRD | 11864612 |
| CSNK2A1 | CK2A1 | CKII | casein kinase 2, alpha 1 polypeptide | - | HPRD,BioGRID | 7794926 |
| CSNK2A2 | CK2A2 | CSNK2A1 | FLJ43934 | casein kinase 2, alpha prime polypeptide | - | HPRD,BioGRID | 7794926 |
| CUL4A | - | cullin 4A | Affinity Capture-MS | BioGRID | 12481031 |
| DAP3 | DAP-3 | DKFZp686G12159 | MGC126058 | MGC126059 | MRP-S29 | MRPS29 | bMRP-10 | death associated protein 3 | - | HPRD,BioGRID | 10903152 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | EGFR interacts with Hsp90. This interaction was modeled on a demonstrated interaction between EGFR from an unspecified species and human Hsp90. | BIND | 12471035 |
| EIF2AK2 | EIF2AK1 | MGC126524 | PKR | PRKR | eukaryotic translation initiation factor 2-alpha kinase 2 | - | HPRD,BioGRID | 11447118 |
| EIF2AK3 | DKFZp781H1925 | HRI | PEK | PERK | WRS | eukaryotic translation initiation factor 2-alpha kinase 3 | - | HPRD,BioGRID | 12446770 |
| EPRS | DKFZp313B047 | EARS | GLUPRORS | PARS | PIG32 | QARS | QPRS | glutamyl-prolyl-tRNA synthetase | - | HPRD,BioGRID | 10913161 |
| ERBB2 | CD340 | HER-2 | HER-2/neu | HER2 | NEU | NGL | TKR1 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | ERBB2 interacts with HSP90. This interaction was modelled on a demonstrated interaction between human ERBB2 and monkey HSP90. | BIND | 15643424 |
| ERBB2 | CD340 | HER-2 | HER-2/neu | HER2 | NEU | NGL | TKR1 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | - | HPRD,BioGRID | 11071886 |
| ERN1 | FLJ30999 | IRE1 | IRE1P | MGC163277 | MGC163279 | endoplasmic reticulum to nucleus signaling 1 | - | HPRD,BioGRID | 12446770 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD,BioGRID | 11911945 |
| FANCA | FA | FA-H | FA1 | FAA | FACA | FAH | FANCH | MGC75158 | Fanconi anemia, complementation group A | Two-hybrid | BioGRID | 14499622 |
| FANCC | FA3 | FAC | FACC | FLJ14675 | Fanconi anemia, complementation group C | Two-hybrid | BioGRID | 14499622 |
| FES | FPS | feline sarcoma oncogene | Reconstituted Complex | BioGRID | 9222609 |
| FKBP5 | FKBP51 | FKBP54 | MGC111006 | P54 | PPIase | Ptg-10 | FK506 binding protein 5 | - | HPRD,BioGRID | 9001212 |12538866 |
| FNIP1 | DKFZp686E18167 | DKFZp781P0215 | KIAA1961 | MGC667 | folliculin interacting protein 1 | Affinity Capture-MS | BioGRID | 17028174 |
| GNA12 | MGC104623 | MGC99644 | NNX3 | RMP | gep | guanine nucleotide binding protein (G protein) alpha 12 | in vitro in vivo Two-hybrid | BioGRID | 11598136 |12117999 |
| GNA12 | MGC104623 | MGC99644 | NNX3 | RMP | gep | guanine nucleotide binding protein (G protein) alpha 12 | - | HPRD | 11598136|12117999 |
| GUCY1B3 | GC-S-beta-1 | GC-SB3 | GUC1B3 | GUCB3 | GUCSB3 | GUCY1B1 | guanylate cyclase 1, soluble, beta 3 | - | HPRD,BioGRID | 12676772 |
| HCFC1 | CFF | HCF-1 | HCF1 | HFC1 | MGC70925 | VCAF | host cell factor C1 (VP16-accessory protein) | Affinity Capture-MS | BioGRID | 12670868 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | - | HPRD,BioGRID | 9079689 |
| HSF1 | HSTF1 | heat shock transcription factor 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9222609 |12621024 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | Two-hybrid | BioGRID | 16169070 |
| HSPA1A | FLJ54303 | FLJ54370 | FLJ54392 | FLJ54408 | FLJ75127 | HSP70-1 | HSP70-1A | HSP70I | HSP72 | HSPA1 | HSPA1B | heat shock 70kDa protein 1A | Reconstituted Complex | BioGRID | 12093808 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | - | HPRD,BioGRID | 9269769 |
| IKBKB | FLJ40509 | IKK-beta | IKK2 | IKKB | MGC131801 | NFKBIKB | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta | - | HPRD | 11864612 |
| IKBKG | AMCBX1 | FIP-3 | FIP3 | Fip3p | IKK-gamma | IP | IP1 | IP2 | IPD2 | NEMO | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma | - | HPRD | 11864612 |
| IRS4 | IRS-4 | PY160 | insulin receptor substrate 4 | Affinity Capture-MS | BioGRID | 11912194 |
| LSM1 | CASM | YJL124C | LSM1 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD,BioGRID | 15231747 |
| MAP3K7 | TAK1 | TGF1a | mitogen-activated protein kinase kinase kinase 7 | - | HPRD | 14743216 |
| MAP3K7IP2 | FLJ21885 | KIAA0733 | TAB2 | mitogen-activated protein kinase kinase kinase 7 interacting protein 2 | - | HPRD | 14743216 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | - | HPRD | 11748628 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | - | HPRD,BioGRID | 12644583 |
| MYOD1 | MYF3 | MYOD | PUM | bHLHc1 | myogenic differentiation 1 | - | HPRD,BioGRID | 1406681 |
| NOS3 | ECNOS | eNOS | nitric oxide synthase 3 (endothelial cell) | - | HPRD,BioGRID | 11744239 |12124224 |
| NPAS2 | FLJ23138 | MGC71151 | MOP4 | PASD4 | bHLHe9 | neuronal PAS domain protein 2 | Reconstituted Complex | BioGRID | 9079689 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | - | HPRD,BioGRID | 8621522 |8645634 |8898375 |9334248 |10066374 |
| NR3C2 | MCR | MGC133092 | MLR | MR | nuclear receptor subfamily 3, group C, member 2 | - | HPRD | 9392437 |
| PDPK1 | MGC20087 | MGC35290 | PDK1 | PRO0461 | 3-phosphoinositide dependent protein kinase-1 | - | HPRD | 11779851 |
| PIM1 | PIM | pim-1 oncogene | - | HPRD,BioGRID | 11237709 |
| PPARA | MGC2237 | MGC2452 | NR1C1 | PPAR | hPPAR | peroxisome proliferator-activated receptor alpha | - | HPRD,BioGRID | 12482853 |
| PPID | CYP-40 | CYPD | MGC33096 | peptidylprolyl isomerase D | - | HPRD | 12145316 |
| PPP5C | FLJ36922 | PP5 | PPP5 | protein phosphatase 5, catalytic subunit | - | HPRD | 10400612 |
| PPP5C | FLJ36922 | PP5 | PPP5 | protein phosphatase 5, catalytic subunit | Hsp90 interacts with PP5. This interaction was modeled on a demonstrated interaction between human Hsp90 and PP5 from an unspecified species. | BIND | 15664193 |
| PTGES3 | P23 | TEBP | cPGES | prostaglandin E synthase 3 (cytosolic) | - | HPRD,BioGRID | 11812147 |12077419 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | Raf-1 interacts with Hsp90. This interaction was modeled on a demonstrated interaction between human Raf-1 and monkey Hsp90. | BIND | 15618521 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | - | HPRD,BioGRID | 8408024 |
| RALBP1 | RIP1 | RLIP1 | RLIP76 | ralA binding protein 1 | Affinity Capture-Western | BioGRID | 12621024 |
| RIPK1 | FLJ39204 | RIP | RIP1 | receptor (TNFRSF)-interacting serine-threonine kinase 1 | - | HPRD | 10744744 |
| RPS3A | FTE1 | MFTL | MGC23240 | ribosomal protein S3A | Two-hybrid | BioGRID | 16169070 |
| RUNX1T1 | AML1T1 | CBFA2T1 | CDR | ETO | MGC2796 | MTG8 | MTG8b | ZMYND2 | runt-related transcription factor 1; translocated to, 1 (cyclin D-related) | - | HPRD | 10076566 |
| SGTA | SGT | alphaSGT | hSGT | small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha | - | HPRD,BioGRID | 12482202 |
| SIM1 | bHLHe14 | single-minded homolog 1 (Drosophila) | Reconstituted Complex | BioGRID | 9020169 |
| SMYD3 | FLJ21080 | MGC104324 | ZMYND1 | ZNFN3A1 | bA74P14.1 | SET and MYND domain containing 3 | SMYD3 interacts with HSP90A. | BIND | 15235609 |
| SMYD3 | FLJ21080 | MGC104324 | ZMYND1 | ZNFN3A1 | bA74P14.1 | SET and MYND domain containing 3 | - | HPRD,BioGRID | 15235609 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD,BioGRID | 1310678 |
| STARD13 | DLC2 | FLJ37385 | GT650 | StAR-related lipid transfer (START) domain containing 13 | Two-hybrid | BioGRID | 14697242 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | - | HPRD,BioGRID | 12559950 |
| STIP1 | HOP | IEF-SSP-3521 | P60 | STI1 | STI1L | stress-induced-phosphoprotein 1 | - | HPRD,BioGRID | 10786835 |
| STK11 | LKB1 | PJS | serine/threonine kinase 11 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12489981 |
| STUB1 | CHIP | HSPABP2 | NY-CO-7 | SDCCAG7 | UBOX1 | STIP1 homology and U-box containing protein 1 | - | HPRD | 11146632|11557750 |
| TERT | EST2 | TCS1 | TP2 | TRT | hEST2 | telomerase reverse transcriptase | - | HPRD,BioGRID | 12586360 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | - | HPRD | 7876093 |
| TOMM34 | HTOM34P | TOM34 | URCC3 | translocase of outer mitochondrial membrane 34 | - | HPRD | 9660753 |
| TOMM70A | FLJ90470 | translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) | - | HPRD | 12526792 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 11507088 |12427754 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | - | HPRD | 14743216 |
| TUBA1A | B-ALPHA-1 | FLJ25113 | LIS3 | TUBA3 | tubulin, alpha 1a | Affinity Capture-Western | BioGRID | 12621024 |
| WASL | DKFZp779G0847 | MGC48327 | N-WASP | NWASP | Wiskott-Aldrich syndrome-like | WASL (N-WASP) interacts with HSPCA (HSP90-alpha). | BIND | 15791211 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ANTIGEN PROCESSING AND PRESENTATION | 89 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG NOD LIKE RECEPTOR SIGNALING PATHWAY | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG PROGESTERONE MEDIATED OOCYTE MATURATION | 86 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER | 89 | 75 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NO1 PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AKT PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GCR PATHWAY | 22 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P53HYPOXIA PATHWAY | 23 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIF PATHWAY | 15 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PPARA PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TEL PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSII PATHWAY | 34 | 27 | All SZGR 2.0 genes in this pathway |
| PID MYC ACTIV PATHWAY | 79 | 62 | All SZGR 2.0 genes in this pathway |
| PID ILK PATHWAY | 45 | 32 | All SZGR 2.0 genes in this pathway |
| PID LKB1 PATHWAY | 47 | 37 | All SZGR 2.0 genes in this pathway |
| PID TELOMERASE PATHWAY | 68 | 48 | All SZGR 2.0 genes in this pathway |
| PID REG GR PATHWAY | 82 | 60 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI PATHWAY | 49 | 40 | All SZGR 2.0 genes in this pathway |
| PID IL2 PI3K PATHWAY | 34 | 27 | All SZGR 2.0 genes in this pathway |
| PID AR TF PATHWAY | 53 | 38 | All SZGR 2.0 genes in this pathway |
| PID AVB3 INTEGRIN PATHWAY | 75 | 53 | All SZGR 2.0 genes in this pathway |
| PID HIF1A PATHWAY | 19 | 12 | All SZGR 2.0 genes in this pathway |
| PID ERBB NETWORK PATHWAY | 15 | 13 | All SZGR 2.0 genes in this pathway |
| PID VEGFR1 PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| PID VEGFR1 2 PATHWAY | 69 | 57 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI AKT PATHWAY | 35 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | 13 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2 | 101 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | 18 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER | 109 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME ENOS ACTIVATION AND REGULATION | 20 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | 66 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | 59 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | 15 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMAPHORIN INTERACTIONS | 68 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G2 G2 M PHASES | 81 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLUENZA LIFE CYCLE | 203 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | 169 | 47 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP | 285 | 181 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS TOP50 UP | 38 | 27 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS DN | 310 | 188 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION UP | 119 | 66 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 UP | 276 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 2 UP | 139 | 83 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 UP | 341 | 197 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS DN | 73 | 51 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP | 164 | 122 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS UP | 214 | 134 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP UP | 265 | 158 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR DN | 75 | 50 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| MARKS ACETYLATED NON HISTONE PROTEINS | 15 | 9 | All SZGR 2.0 genes in this pathway |
| SEIDEN ONCOGENESIS BY MET | 88 | 53 | All SZGR 2.0 genes in this pathway |
| SCHEIDEREIT IKK INTERACTING PROTEINS | 58 | 45 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| COLLIS PRKDC SUBSTRATES | 20 | 15 | All SZGR 2.0 genes in this pathway |
| MENSSEN MYC TARGETS | 53 | 35 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION DN | 187 | 122 | All SZGR 2.0 genes in this pathway |
| UEDA PERIFERAL CLOCK | 169 | 111 | All SZGR 2.0 genes in this pathway |
| POMEROY MEDULLOBLASTOMA DESMOPLASIC VS CLASSIC UP | 62 | 38 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| SHIPP DLBCL VS FOLLICULAR LYMPHOMA UP | 45 | 30 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE DN | 245 | 154 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS UP | 151 | 103 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| APRELIKOVA BRCA1 TARGETS | 49 | 33 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS DN | 215 | 132 | All SZGR 2.0 genes in this pathway |
| ZHANG PROLIFERATING VS QUIESCENT | 51 | 41 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT DN | 165 | 106 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C1 | 72 | 45 | All SZGR 2.0 genes in this pathway |
| WENG POR TARGETS GLOBAL DN | 23 | 14 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS UP | 113 | 71 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| CHENG RESPONSE TO NICKEL ACETATE | 45 | 29 | All SZGR 2.0 genes in this pathway |
| WENG POR TARGETS LIVER DN | 20 | 11 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 2 | 473 | 224 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL UP | 125 | 68 | All SZGR 2.0 genes in this pathway |
| BRUECKNER TARGETS OF MIRLET7A3 DN | 78 | 49 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| CHAUHAN RESPONSE TO METHOXYESTRADIOL UP | 51 | 32 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN MYELOMA VS MATURE B LYMPHOCYTE | 101 | 76 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER UP | 83 | 63 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN | 374 | 217 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| GUO TARGETS OF IRS1 AND IRS2 | 98 | 67 | All SZGR 2.0 genes in this pathway |
| FU INTERACT WITH ALKBH8 | 13 | 8 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-361 | 46 | 53 | 1A,m8 | hsa-miR-361brain | UUAUCAGAAUCUCCAGGGGUAC |
| miR-495 | 82 | 88 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.