Gene Page: APOB
Summary ?
| GeneID | 338 |
| Symbol | APOB |
| Synonyms | FLDB|LDLCQ4|apoB-100|apoB-48 |
| Description | apolipoprotein B |
| Reference | MIM:107730|HGNC:HGNC:603|Ensembl:ENSG00000084674|HPRD:00133|Vega:OTTHUMG00000090785 |
| Gene type | protein-coding |
| Map location | 2p24-p23 |
| Pascal p-value | 0.689 |
| Fetal beta | -0.366 |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
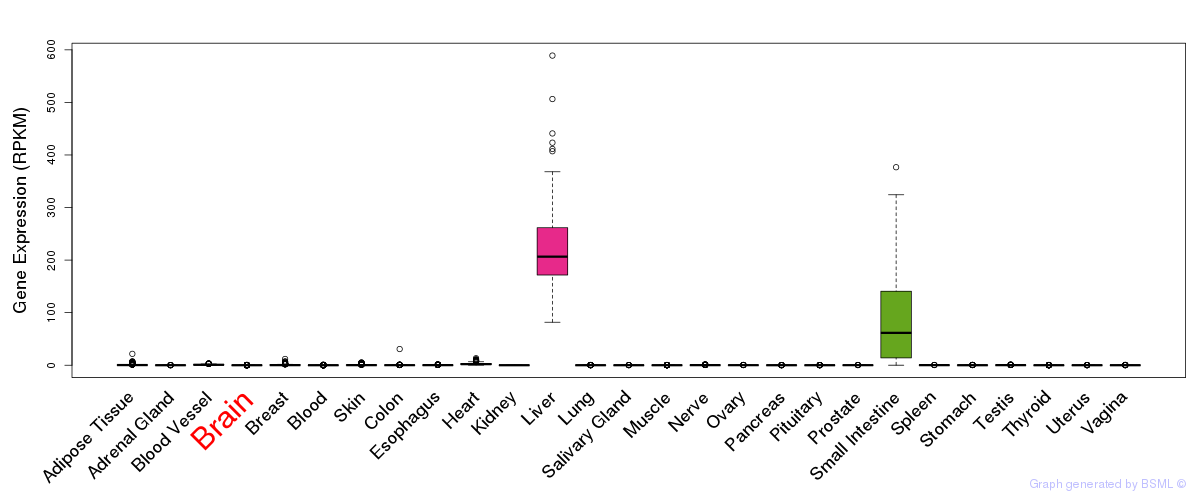
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| VAMP5 | 0.76 | 0.84 |
| C1orf54 | 0.74 | 0.82 |
| GNG11 | 0.69 | 0.70 |
| C1orf61 | 0.69 | 0.72 |
| METRN | 0.69 | 0.86 |
| AL138743.2 | 0.68 | 0.69 |
| IFI27 | 0.68 | 0.86 |
| AF347015.21 | 0.67 | 0.81 |
| S100A13 | 0.67 | 0.80 |
| IFI27L2 | 0.67 | 0.73 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ADD1 | -0.64 | -0.79 |
| VPS52 | -0.63 | -0.57 |
| DDX42 | -0.63 | -0.77 |
| AC119673.2 | -0.62 | -0.67 |
| RABGAP1 | -0.62 | -0.75 |
| RABGEF1 | -0.61 | -0.78 |
| EIF4ENIF1 | -0.61 | -0.81 |
| CSRP2BP | -0.61 | -0.71 |
| HNRNPH2 | -0.61 | -0.67 |
| ARFGAP2 | -0.61 | -0.66 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BGN | DSPG1 | PG-S1 | PGI | SLRR1A | biglycan | - | HPRD,BioGRID | 12070165 |
| CALCR | CRT | CTR | CTR1 | calcitonin receptor | Affinity Capture-RNA Affinity Capture-Western Co-fractionation | BioGRID | 9694898 |12397072 |
| CALR | CRT | FLJ26680 | RO | SSA | cC1qR | calreticulin | - | HPRD,BioGRID | 10513896 |12397072 |
| CANX | CNX | FLJ26570 | IP90 | P90 | calnexin | Affinity Capture-Western | BioGRID | 14498830 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | - | HPRD | 11333259 |
| HSP90B1 | ECGP | GP96 | GRP94 | TRA1 | heat shock protein 90kDa beta (Grp94), member 1 | - | HPRD,BioGRID | 12397072 |
| HSPA1A | FLJ54303 | FLJ54370 | FLJ54392 | FLJ54408 | FLJ75127 | HSP70-1 | HSP70-1A | HSP70I | HSP72 | HSPA1 | HSPA1B | heat shock 70kDa protein 1A | - | HPRD | 9252351 |
| HSPA5 | BIP | FLJ26106 | GRP78 | MIF2 | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) | - | HPRD | 12397072 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | - | HPRD | 9694898 |
| LDLR | FH | FHC | low density lipoprotein receptor | - | HPRD | 12031600 |
| LIPC | HDLCQ12 | HL | HTGL | LIPH | lipase, hepatic | - | HPRD,BioGRID | 9685400 |
| LRP2 | DBS | gp330 | low density lipoprotein-related protein 2 | - | HPRD | 10330424 |
| MTTP | ABL | MGC149819 | MGC149820 | MTP | microsomal triglyceride transfer protein | - | HPRD,BioGRID | 9915855 |11358959 |
| PDIA4 | ERP70 | ERP72 | protein disulfide isomerase family A, member 4 | - | HPRD,BioGRID | 9694898 |12397072 |
| PPIB | CYP-S1 | CYPB | MGC14109 | MGC2224 | SCYLP | peptidylprolyl isomerase B (cyclophilin B) | - | HPRD,BioGRID | 12397072 |
| SEC61A1 | HSEC61 | SEC61 | SEC61A | Sec61 alpha 1 subunit (S. cerevisiae) | Affinity Capture-Western | BioGRID | 9565615 |
| SEC61A2 | FLJ10578 | Sec61 alpha 2 subunit (S. cerevisiae) | - | HPRD | 9565615 |
| SEC61B | - | Sec61 beta subunit | - | HPRD,BioGRID | 9565615 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID AMB2 NEUTROPHILS PATHWAY | 41 | 32 | All SZGR 2.0 genes in this pathway |
| PID HNF3A PATHWAY | 44 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | 91 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET HOMEOSTASIS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET SENSITIZATION BY LDL | 16 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | 46 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME LIPOPROTEIN METABOLISM | 28 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | 16 | 15 | All SZGR 2.0 genes in this pathway |
| SMIRNOV CIRCULATING ENDOTHELIOCYTES IN CANCER UP | 158 | 103 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| IIZUKA LIVER CANCER PROGRESSION L0 L1 UP | 17 | 12 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| URS ADIPOCYTE DIFFERENTIATION UP | 74 | 51 | All SZGR 2.0 genes in this pathway |
| SU LIVER | 55 | 32 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX DN | 88 | 58 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS DN | 96 | 71 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOZOA | 114 | 77 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING UP | 93 | 62 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |