Gene Page: ICAM3
Summary ?
| GeneID | 3385 |
| Symbol | ICAM3 |
| Synonyms | CD50|CDW50|ICAM-R |
| Description | intercellular adhesion molecule 3 |
| Reference | MIM:146631|HGNC:HGNC:5346|Ensembl:ENSG00000076662|HPRD:00889|Vega:OTTHUMG00000180410 |
| Gene type | protein-coding |
| Map location | 19p13.2 |
| Pascal p-value | 0.19 |
| Sherlock p-value | 0.542 |
| Fetal beta | -0.519 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellum Cortex Hippocampus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17126555 | 19 | 10445516 | ICAM3 | 3.17E-4 | 0.006 | 0.183 | DMG:Montano_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs3176767 | chr19 | 10449750 | ICAM3 | 3385 | 5.825E-6 | cis | ||
| rs17151821 | chr7 | 8628490 | ICAM3 | 3385 | 0.01 | trans | ||
| rs16946889 | chr15 | 92964427 | ICAM3 | 3385 | 0.06 | trans | ||
| rs10853963 | chr19 | 2909032 | ICAM3 | 3385 | 0.06 | trans | ||
| rs4807364 | chr19 | 2923262 | ICAM3 | 3385 | 0.08 | trans | ||
| rs3176767 | chr19 | 10449750 | ICAM3 | 3385 | 9.526E-4 | trans | ||
| rs2304237 | 19 | 10446568 | ICAM3 | ENSG00000076662.5 | 1.939E-9 | 0 | 3931 | gtex_brain_ba24 |
| rs1058154 | 19 | 10446897 | ICAM3 | ENSG00000076662.5 | 2.126E-9 | 0 | 3602 | gtex_brain_ba24 |
| rs74312588 | 19 | 10446900 | ICAM3 | ENSG00000076662.5 | 5.498E-9 | 0 | 3599 | gtex_brain_ba24 |
| rs3176769 | 19 | 10447034 | ICAM3 | ENSG00000076662.5 | 2.316E-9 | 0 | 3465 | gtex_brain_ba24 |
| rs10408916 | 19 | 10447770 | ICAM3 | ENSG00000076662.5 | 4.138E-9 | 0 | 2729 | gtex_brain_ba24 |
| rs10403396 | 19 | 10447819 | ICAM3 | ENSG00000076662.5 | 4.153E-9 | 0 | 2680 | gtex_brain_ba24 |
| rs7253912 | 19 | 10448555 | ICAM3 | ENSG00000076662.5 | 1.075E-8 | 0 | 1944 | gtex_brain_ba24 |
| rs7250656 | 19 | 10448566 | ICAM3 | ENSG00000076662.5 | 1.09E-8 | 0 | 1933 | gtex_brain_ba24 |
| rs7254041 | 19 | 10448636 | ICAM3 | ENSG00000076662.5 | 1.192E-8 | 0 | 1863 | gtex_brain_ba24 |
| rs7250300 | 19 | 10448660 | ICAM3 | ENSG00000076662.5 | 1.243E-8 | 0 | 1839 | gtex_brain_ba24 |
| rs7254330 | 19 | 10448876 | ICAM3 | ENSG00000076662.5 | 1.687E-8 | 0 | 1623 | gtex_brain_ba24 |
| rs7249914 | 19 | 10449009 | ICAM3 | ENSG00000076662.5 | 2.062E-8 | 0 | 1490 | gtex_brain_ba24 |
| rs7257871 | 19 | 10449252 | ICAM3 | ENSG00000076662.5 | 3.017E-8 | 0 | 1247 | gtex_brain_ba24 |
| rs7258015 | 19 | 10449358 | ICAM3 | ENSG00000076662.5 | 3.017E-8 | 0 | 1141 | gtex_brain_ba24 |
| rs3176768 | 19 | 10449665 | ICAM3 | ENSG00000076662.5 | 2.629E-9 | 0 | 834 | gtex_brain_ba24 |
| rs3176767 | 19 | 10449751 | ICAM3 | ENSG00000076662.5 | 3.017E-8 | 0 | 748 | gtex_brain_ba24 |
| rs3176766 | 19 | 10449778 | ICAM3 | ENSG00000076662.5 | 3.006E-8 | 0 | 721 | gtex_brain_ba24 |
| rs2304237 | 19 | 10446568 | ICAM3 | ENSG00000076662.5 | 2.657E-8 | 0 | 3931 | gtex_brain_putamen_basal |
| rs1058154 | 19 | 10446897 | ICAM3 | ENSG00000076662.5 | 2.867E-8 | 0 | 3602 | gtex_brain_putamen_basal |
| rs74312588 | 19 | 10446900 | ICAM3 | ENSG00000076662.5 | 2.53E-8 | 0 | 3599 | gtex_brain_putamen_basal |
| rs3176769 | 19 | 10447034 | ICAM3 | ENSG00000076662.5 | 2.994E-8 | 0 | 3465 | gtex_brain_putamen_basal |
| rs10408916 | 19 | 10447770 | ICAM3 | ENSG00000076662.5 | 4.395E-8 | 0 | 2729 | gtex_brain_putamen_basal |
| rs10403396 | 19 | 10447819 | ICAM3 | ENSG00000076662.5 | 3.198E-8 | 0 | 2680 | gtex_brain_putamen_basal |
| rs7253912 | 19 | 10448555 | ICAM3 | ENSG00000076662.5 | 6.458E-8 | 0 | 1944 | gtex_brain_putamen_basal |
| rs7250656 | 19 | 10448566 | ICAM3 | ENSG00000076662.5 | 6.479E-8 | 0 | 1933 | gtex_brain_putamen_basal |
| rs7254041 | 19 | 10448636 | ICAM3 | ENSG00000076662.5 | 6.764E-8 | 0 | 1863 | gtex_brain_putamen_basal |
| rs7250300 | 19 | 10448660 | ICAM3 | ENSG00000076662.5 | 6.931E-8 | 0 | 1839 | gtex_brain_putamen_basal |
| rs7254330 | 19 | 10448876 | ICAM3 | ENSG00000076662.5 | 8.077E-8 | 0 | 1623 | gtex_brain_putamen_basal |
| rs7249914 | 19 | 10449009 | ICAM3 | ENSG00000076662.5 | 8.924E-8 | 0 | 1490 | gtex_brain_putamen_basal |
| rs7257871 | 19 | 10449252 | ICAM3 | ENSG00000076662.5 | 1.08E-7 | 0 | 1247 | gtex_brain_putamen_basal |
| rs7258015 | 19 | 10449358 | ICAM3 | ENSG00000076662.5 | 1.08E-7 | 0 | 1141 | gtex_brain_putamen_basal |
| rs3176768 | 19 | 10449665 | ICAM3 | ENSG00000076662.5 | 2.548E-8 | 0 | 834 | gtex_brain_putamen_basal |
| rs3176767 | 19 | 10449751 | ICAM3 | ENSG00000076662.5 | 1.08E-7 | 0 | 748 | gtex_brain_putamen_basal |
| rs3176766 | 19 | 10449778 | ICAM3 | ENSG00000076662.5 | 1.081E-7 | 0 | 721 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
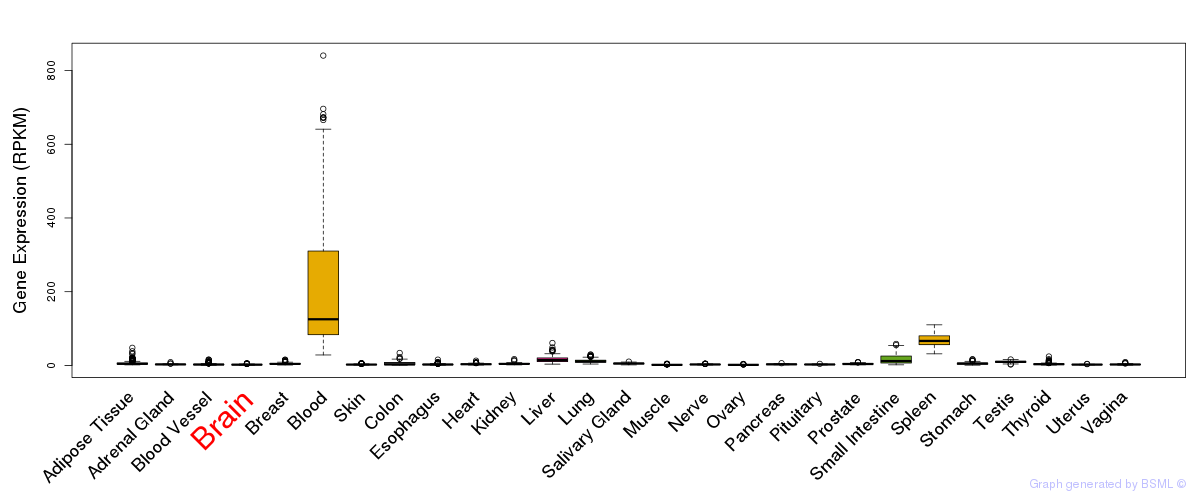
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN2 PATHWAY | 29 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | 70 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN CELL SURFACE INTERACTIONS | 79 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| LIU CMYB TARGETS UP | 165 | 106 | All SZGR 2.0 genes in this pathway |
| LIU VMYB TARGETS UP | 127 | 78 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE DN | 54 | 38 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| DUNNE TARGETS OF AML1 MTG8 FUSION UP | 52 | 32 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL DN | 186 | 107 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER HIGH RECURRENCE | 49 | 31 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE UP | 134 | 93 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP | 309 | 199 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| ADDYA ERYTHROID DIFFERENTIATION BY HEMIN | 73 | 47 | All SZGR 2.0 genes in this pathway |
| XU RESPONSE TO TRETINOIN AND NSC682994 UP | 17 | 13 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING | 246 | 152 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN | 245 | 150 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| KESHELAVA MULTIPLE DRUG RESISTANCE | 88 | 56 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA MESENCHYMAL | 216 | 130 | All SZGR 2.0 genes in this pathway |