Gene Page: FAS
Summary ?
| GeneID | 355 |
| Symbol | FAS |
| Synonyms | ALPS1A|APO-1|APT1|CD95|FAS1|FASTM|TNFRSF6 |
| Description | Fas cell surface death receptor |
| Reference | MIM:134637|HGNC:HGNC:11920|Ensembl:ENSG00000026103|HPRD:00609|Vega:OTTHUMG00000018701 |
| Gene type | protein-coding |
| Map location | 10q24.1 |
| Pascal p-value | 0.654 |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cortex Frontal Cortex BA9 Hippocampus Nucleus accumbens basal ganglia Putamen basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 2.86 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizotypy,schizophrenias,schizotypal | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0203 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7082101 | 10 | 90741615 | FAS | ENSG00000026103.15 | 3.476E-11 | 0 | -8799 | gtex_brain_ba24 |
| rs7069841 | 10 | 90741955 | FAS | ENSG00000026103.15 | 3.456E-11 | 0 | -8459 | gtex_brain_ba24 |
| rs11202918 | 10 | 90742008 | FAS | ENSG00000026103.15 | 7.81E-7 | 0 | -8406 | gtex_brain_ba24 |
| rs7082720 | 10 | 90742049 | FAS | ENSG00000026103.15 | 3.483E-11 | 0 | -8365 | gtex_brain_ba24 |
| rs7910660 | 10 | 90743197 | FAS | ENSG00000026103.15 | 3.442E-11 | 0 | -7217 | gtex_brain_ba24 |
| rs7910825 | 10 | 90743341 | FAS | ENSG00000026103.15 | 3.442E-11 | 0 | -7073 | gtex_brain_ba24 |
| rs3903454 | 10 | 90743434 | FAS | ENSG00000026103.15 | 8.059E-7 | 0 | -6980 | gtex_brain_ba24 |
| rs1926201 | 10 | 90746360 | FAS | ENSG00000026103.15 | 3.442E-11 | 0 | -4054 | gtex_brain_ba24 |
| rs1926200 | 10 | 90746533 | FAS | ENSG00000026103.15 | 3.442E-11 | 0 | -3881 | gtex_brain_ba24 |
| rs1926199 | 10 | 90746660 | FAS | ENSG00000026103.15 | 3.442E-11 | 0 | -3754 | gtex_brain_ba24 |
| rs4934434 | 10 | 90747169 | FAS | ENSG00000026103.15 | 1.052E-6 | 0 | -3245 | gtex_brain_ba24 |
| rs7914490 | 10 | 90747447 | FAS | ENSG00000026103.15 | 3.434E-11 | 0 | -2967 | gtex_brain_ba24 |
| rs3758485 | 10 | 90747847 | FAS | ENSG00000026103.15 | 1.302E-6 | 0 | -2567 | gtex_brain_ba24 |
| rs978522 | 10 | 90748338 | FAS | ENSG00000026103.15 | 1.565E-6 | 0 | -2076 | gtex_brain_ba24 |
| rs1926197 | 10 | 90748626 | FAS | ENSG00000026103.15 | 3.43E-11 | 0 | -1788 | gtex_brain_ba24 |
| rs1800682 | 10 | 90749963 | FAS | ENSG00000026103.15 | 3.441E-11 | 0 | -451 | gtex_brain_ba24 |
| rs12775501 | 10 | 90750909 | FAS | ENSG00000026103.15 | 3.337E-8 | 0 | 495 | gtex_brain_ba24 |
| rs3740286 | 10 | 90751340 | FAS | ENSG00000026103.15 | 2.97E-8 | 0 | 926 | gtex_brain_ba24 |
| rs1324551 | 10 | 90751516 | FAS | ENSG00000026103.15 | 6.852E-13 | 0 | 1102 | gtex_brain_ba24 |
| rs6586163 | 10 | 90752018 | FAS | ENSG00000026103.15 | 1.116E-12 | 0 | 1604 | gtex_brain_ba24 |
| rs6586164 | 10 | 90752031 | FAS | ENSG00000026103.15 | 1.116E-12 | 0 | 1617 | gtex_brain_ba24 |
| rs3824730 | 10 | 90752128 | FAS | ENSG00000026103.15 | 2.887E-8 | 0 | 1714 | gtex_brain_ba24 |
| rs3824729 | 10 | 90752616 | FAS | ENSG00000026103.15 | 1.116E-12 | 0 | 2202 | gtex_brain_ba24 |
| rs10887877 | 10 | 90753038 | FAS | ENSG00000026103.15 | 1.116E-12 | 0 | 2624 | gtex_brain_ba24 |
| rs12359362 | 10 | 90753145 | FAS | ENSG00000026103.15 | 2.788E-8 | 0 | 2731 | gtex_brain_ba24 |
| rs7097572 | 10 | 90753260 | FAS | ENSG00000026103.15 | 1.116E-12 | 0 | 2846 | gtex_brain_ba24 |
| rs5786849 | 10 | 90753549 | FAS | ENSG00000026103.15 | 1.116E-12 | 0 | 3135 | gtex_brain_ba24 |
| rs1926196 | 10 | 90753748 | FAS | ENSG00000026103.15 | 1.116E-12 | 0 | 3334 | gtex_brain_ba24 |
| rs1926195 | 10 | 90753758 | FAS | ENSG00000026103.15 | 1.249E-12 | 0 | 3344 | gtex_brain_ba24 |
| rs2031610 | 10 | 90754487 | FAS | ENSG00000026103.15 | 2.724E-8 | 0 | 4073 | gtex_brain_ba24 |
| rs7897395 | 10 | 90754607 | FAS | ENSG00000026103.15 | 2.718E-12 | 0 | 4193 | gtex_brain_ba24 |
| rs7069061 | 10 | 90754909 | FAS | ENSG00000026103.15 | 1.271E-12 | 0 | 4495 | gtex_brain_ba24 |
| rs7072828 | 10 | 90754953 | FAS | ENSG00000026103.15 | 1.306E-12 | 0 | 4539 | gtex_brain_ba24 |
| rs6586165 | 10 | 90755056 | FAS | ENSG00000026103.15 | 1.372E-12 | 0 | 4642 | gtex_brain_ba24 |
| rs6586166 | 10 | 90756065 | FAS | ENSG00000026103.15 | 4.382E-12 | 0 | 5651 | gtex_brain_ba24 |
| rs6586167 | 10 | 90756146 | FAS | ENSG00000026103.15 | 4.729E-12 | 0 | 5732 | gtex_brain_ba24 |
| rs2147420 | 10 | 90759613 | FAS | ENSG00000026103.15 | 1.403E-8 | 0 | 9199 | gtex_brain_ba24 |
| rs4406737 | 10 | 90759724 | FAS | ENSG00000026103.15 | 1.403E-8 | 0 | 9310 | gtex_brain_ba24 |
| rs2148287 | 10 | 90759772 | FAS | ENSG00000026103.15 | 2.705E-8 | 0 | 9358 | gtex_brain_ba24 |
| rs4406738 | 10 | 90760012 | FAS | ENSG00000026103.15 | 1.403E-8 | 0 | 9598 | gtex_brain_ba24 |
| rs2182408 | 10 | 90760107 | FAS | ENSG00000026103.15 | 1.408E-8 | 0 | 9693 | gtex_brain_ba24 |
| rs1926194 | 10 | 90760318 | FAS | ENSG00000026103.15 | 8.28E-9 | 0 | 9904 | gtex_brain_ba24 |
| rs1926193 | 10 | 90760606 | FAS | ENSG00000026103.15 | 9.36E-9 | 0 | 10192 | gtex_brain_ba24 |
| rs1926192 | 10 | 90760620 | FAS | ENSG00000026103.15 | 8.28E-9 | 0 | 10206 | gtex_brain_ba24 |
| rs9658720 | 10 | 90760669 | FAS | ENSG00000026103.15 | 8.527E-9 | 0 | 10255 | gtex_brain_ba24 |
| rs7069750 | 10 | 90762376 | FAS | ENSG00000026103.15 | 1.181E-8 | 0 | 11962 | gtex_brain_ba24 |
| rs9658742 | 10 | 90764891 | FAS | ENSG00000026103.15 | 2.159E-7 | 0 | 14477 | gtex_brain_ba24 |
| rs7901656 | 10 | 90766213 | FAS | ENSG00000026103.15 | 1.323E-6 | 0 | 15799 | gtex_brain_ba24 |
| rs7082101 | 10 | 90741615 | FAS | ENSG00000026103.15 | 6.281E-9 | 0 | -8799 | gtex_brain_putamen_basal |
| rs7069841 | 10 | 90741955 | FAS | ENSG00000026103.15 | 6.269E-9 | 0 | -8459 | gtex_brain_putamen_basal |
| rs11202918 | 10 | 90742008 | FAS | ENSG00000026103.15 | 6.534E-7 | 0 | -8406 | gtex_brain_putamen_basal |
| rs7082720 | 10 | 90742049 | FAS | ENSG00000026103.15 | 6.259E-9 | 0 | -8365 | gtex_brain_putamen_basal |
| rs7910660 | 10 | 90743197 | FAS | ENSG00000026103.15 | 6.247E-9 | 0 | -7217 | gtex_brain_putamen_basal |
| rs7910825 | 10 | 90743341 | FAS | ENSG00000026103.15 | 6.236E-9 | 0 | -7073 | gtex_brain_putamen_basal |
| rs3903454 | 10 | 90743434 | FAS | ENSG00000026103.15 | 6.643E-7 | 0 | -6980 | gtex_brain_putamen_basal |
| rs1926201 | 10 | 90746360 | FAS | ENSG00000026103.15 | 6.261E-9 | 0 | -4054 | gtex_brain_putamen_basal |
| rs1926200 | 10 | 90746533 | FAS | ENSG00000026103.15 | 6.261E-9 | 0 | -3881 | gtex_brain_putamen_basal |
| rs1926199 | 10 | 90746660 | FAS | ENSG00000026103.15 | 6.261E-9 | 0 | -3754 | gtex_brain_putamen_basal |
| rs4934434 | 10 | 90747169 | FAS | ENSG00000026103.15 | 6.639E-7 | 0 | -3245 | gtex_brain_putamen_basal |
| rs7914490 | 10 | 90747447 | FAS | ENSG00000026103.15 | 6.252E-9 | 0 | -2967 | gtex_brain_putamen_basal |
| rs3758485 | 10 | 90747847 | FAS | ENSG00000026103.15 | 6.635E-7 | 0 | -2567 | gtex_brain_putamen_basal |
| rs978522 | 10 | 90748338 | FAS | ENSG00000026103.15 | 6.631E-7 | 0 | -2076 | gtex_brain_putamen_basal |
| rs1926197 | 10 | 90748626 | FAS | ENSG00000026103.15 | 6.251E-9 | 0 | -1788 | gtex_brain_putamen_basal |
| rs1800682 | 10 | 90749963 | FAS | ENSG00000026103.15 | 6.24E-9 | 0 | -451 | gtex_brain_putamen_basal |
| rs12775501 | 10 | 90750909 | FAS | ENSG00000026103.15 | 3.131E-13 | 0 | 495 | gtex_brain_putamen_basal |
| rs3740286 | 10 | 90751340 | FAS | ENSG00000026103.15 | 2.882E-13 | 0 | 926 | gtex_brain_putamen_basal |
| rs4064 | 10 | 90751380 | FAS | ENSG00000026103.15 | 1.069E-6 | 0 | 966 | gtex_brain_putamen_basal |
| rs1324551 | 10 | 90751516 | FAS | ENSG00000026103.15 | 1.886E-14 | 0 | 1102 | gtex_brain_putamen_basal |
| rs10509561 | 10 | 90751912 | FAS | ENSG00000026103.15 | 1.071E-6 | 0 | 1498 | gtex_brain_putamen_basal |
| rs6586163 | 10 | 90752018 | FAS | ENSG00000026103.15 | 1.621E-14 | 0 | 1604 | gtex_brain_putamen_basal |
| rs6586164 | 10 | 90752031 | FAS | ENSG00000026103.15 | 1.621E-14 | 0 | 1617 | gtex_brain_putamen_basal |
| rs3824730 | 10 | 90752128 | FAS | ENSG00000026103.15 | 2.779E-13 | 0 | 1714 | gtex_brain_putamen_basal |
| rs3824729 | 10 | 90752616 | FAS | ENSG00000026103.15 | 1.589E-14 | 0 | 2202 | gtex_brain_putamen_basal |
| rs10887877 | 10 | 90753038 | FAS | ENSG00000026103.15 | 1.562E-14 | 0 | 2624 | gtex_brain_putamen_basal |
| rs12359362 | 10 | 90753145 | FAS | ENSG00000026103.15 | 2.675E-13 | 0 | 2731 | gtex_brain_putamen_basal |
| rs7097572 | 10 | 90753260 | FAS | ENSG00000026103.15 | 1.546E-14 | 0 | 2846 | gtex_brain_putamen_basal |
| rs5786849 | 10 | 90753549 | FAS | ENSG00000026103.15 | 1.531E-14 | 0 | 3135 | gtex_brain_putamen_basal |
| rs1926196 | 10 | 90753748 | FAS | ENSG00000026103.15 | 1.462E-15 | 0 | 3334 | gtex_brain_putamen_basal |
| rs1926195 | 10 | 90753758 | FAS | ENSG00000026103.15 | 1.535E-14 | 0 | 3344 | gtex_brain_putamen_basal |
| rs2031610 | 10 | 90754487 | FAS | ENSG00000026103.15 | 2.64E-13 | 0 | 4073 | gtex_brain_putamen_basal |
| rs7897395 | 10 | 90754607 | FAS | ENSG00000026103.15 | 3.698E-14 | 0 | 4193 | gtex_brain_putamen_basal |
| rs7069061 | 10 | 90754909 | FAS | ENSG00000026103.15 | 1.115E-14 | 0 | 4495 | gtex_brain_putamen_basal |
| rs7072828 | 10 | 90754953 | FAS | ENSG00000026103.15 | 1.725E-14 | 0 | 4539 | gtex_brain_putamen_basal |
| rs6586165 | 10 | 90755056 | FAS | ENSG00000026103.15 | 1.64E-14 | 0 | 4642 | gtex_brain_putamen_basal |
| rs9658686 | 10 | 90755600 | FAS | ENSG00000026103.15 | 1.235E-6 | 0 | 5186 | gtex_brain_putamen_basal |
| rs6586166 | 10 | 90756065 | FAS | ENSG00000026103.15 | 2.043E-14 | 0 | 5651 | gtex_brain_putamen_basal |
| rs6586167 | 10 | 90756146 | FAS | ENSG00000026103.15 | 2.088E-14 | 0 | 5732 | gtex_brain_putamen_basal |
| rs1571011 | 10 | 90757787 | FAS | ENSG00000026103.15 | 4.651E-9 | 0 | 7373 | gtex_brain_putamen_basal |
| rs1571013 | 10 | 90758349 | FAS | ENSG00000026103.15 | 4.704E-9 | 0 | 7935 | gtex_brain_putamen_basal |
| rs7911752 | 10 | 90758588 | FAS | ENSG00000026103.15 | 4.704E-9 | 0 | 8174 | gtex_brain_putamen_basal |
| rs2147420 | 10 | 90759613 | FAS | ENSG00000026103.15 | 1.581E-10 | 0 | 9199 | gtex_brain_putamen_basal |
| rs4406737 | 10 | 90759724 | FAS | ENSG00000026103.15 | 1.581E-10 | 0 | 9310 | gtex_brain_putamen_basal |
| rs2148287 | 10 | 90759772 | FAS | ENSG00000026103.15 | 1.575E-10 | 0 | 9358 | gtex_brain_putamen_basal |
| rs2148286 | 10 | 90759924 | FAS | ENSG00000026103.15 | 4.704E-9 | 0 | 9510 | gtex_brain_putamen_basal |
| rs4406738 | 10 | 90760012 | FAS | ENSG00000026103.15 | 8.894E-11 | 0 | 9598 | gtex_brain_putamen_basal |
| rs2182408 | 10 | 90760107 | FAS | ENSG00000026103.15 | 1.582E-10 | 0 | 9693 | gtex_brain_putamen_basal |
| rs1926194 | 10 | 90760318 | FAS | ENSG00000026103.15 | 1.339E-10 | 0 | 9904 | gtex_brain_putamen_basal |
| rs1926193 | 10 | 90760606 | FAS | ENSG00000026103.15 | 1.405E-10 | 0 | 10192 | gtex_brain_putamen_basal |
| rs1926192 | 10 | 90760620 | FAS | ENSG00000026103.15 | 1.339E-10 | 0 | 10206 | gtex_brain_putamen_basal |
| rs1926191 | 10 | 90760651 | FAS | ENSG00000026103.15 | 4.386E-9 | 0 | 10237 | gtex_brain_putamen_basal |
| rs9658720 | 10 | 90760669 | FAS | ENSG00000026103.15 | 2.214E-10 | 0 | 10255 | gtex_brain_putamen_basal |
| rs7069750 | 10 | 90762376 | FAS | ENSG00000026103.15 | 1.059E-10 | 0 | 11962 | gtex_brain_putamen_basal |
| rs2296603 | 10 | 90763127 | FAS | ENSG00000026103.15 | 4.702E-9 | 0 | 12713 | gtex_brain_putamen_basal |
| rs9658741 | 10 | 90764739 | FAS | ENSG00000026103.15 | 1.525E-8 | 0 | 14325 | gtex_brain_putamen_basal |
| rs9658742 | 10 | 90764891 | FAS | ENSG00000026103.15 | 7.475E-10 | 0 | 14477 | gtex_brain_putamen_basal |
| rs7901656 | 10 | 90766213 | FAS | ENSG00000026103.15 | 9.08E-10 | 0 | 15799 | gtex_brain_putamen_basal |
| rs2031612 | 10 | 90766980 | FAS | ENSG00000026103.15 | 7.849E-8 | 0 | 16566 | gtex_brain_putamen_basal |
| rs7910435 | 10 | 90768240 | FAS | ENSG00000026103.15 | 3.392E-8 | 0 | 17826 | gtex_brain_putamen_basal |
| rs3781202 | 10 | 90769464 | FAS | ENSG00000026103.15 | 3.424E-8 | 0 | 19050 | gtex_brain_putamen_basal |
| rs397729759 | 10 | 90770084 | FAS | ENSG00000026103.15 | 1.656E-8 | 0 | 19670 | gtex_brain_putamen_basal |
| rs200489672 | 10 | 90770087 | FAS | ENSG00000026103.15 | 2.763E-8 | 0 | 19673 | gtex_brain_putamen_basal |
| rs2296600 | 10 | 90770439 | FAS | ENSG00000026103.15 | 7.876E-8 | 0 | 20025 | gtex_brain_putamen_basal |
| rs3752986 | 10 | 90772781 | FAS | ENSG00000026103.15 | 3.424E-8 | 0 | 22367 | gtex_brain_putamen_basal |
| rs3752985 | 10 | 90772834 | FAS | ENSG00000026103.15 | 1.629E-7 | 0 | 22420 | gtex_brain_putamen_basal |
| rs1571019 | 10 | 90773303 | FAS | ENSG00000026103.15 | 1.178E-7 | 0 | 22889 | gtex_brain_putamen_basal |
| rs1977389 | 10 | 90773494 | FAS | ENSG00000026103.15 | 7.876E-8 | 0 | 23080 | gtex_brain_putamen_basal |
| rs2862833 | 10 | 90775629 | FAS | ENSG00000026103.15 | 1.629E-7 | 0 | 25215 | gtex_brain_putamen_basal |
| rs35150762 | 10 | 90777624 | FAS | ENSG00000026103.15 | 1.629E-7 | 0 | 27210 | gtex_brain_putamen_basal |
| rs1810763 | 10 | 90778829 | FAS | ENSG00000026103.15 | 1.629E-7 | 0 | 28415 | gtex_brain_putamen_basal |
| rs913043 | 10 | 90778967 | FAS | ENSG00000026103.15 | 3.424E-8 | 0 | 28553 | gtex_brain_putamen_basal |
| rs7096473 | 10 | 90780214 | FAS | ENSG00000026103.15 | 7.855E-8 | 0 | 29800 | gtex_brain_putamen_basal |
| rs7089558 | 10 | 90781152 | FAS | ENSG00000026103.15 | 1.49E-7 | 0 | 30738 | gtex_brain_putamen_basal |
| rs10887882 | 10 | 90782922 | FAS | ENSG00000026103.15 | 3.424E-8 | 0 | 32508 | gtex_brain_putamen_basal |
| rs10887883 | 10 | 90782973 | FAS | ENSG00000026103.15 | 3.424E-8 | 0 | 32559 | gtex_brain_putamen_basal |
| rs12775232 | 10 | 90784418 | FAS | ENSG00000026103.15 | 7.988E-8 | 0 | 34004 | gtex_brain_putamen_basal |
| rs7095466 | 10 | 90784755 | FAS | ENSG00000026103.15 | 3.508E-8 | 0 | 34341 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
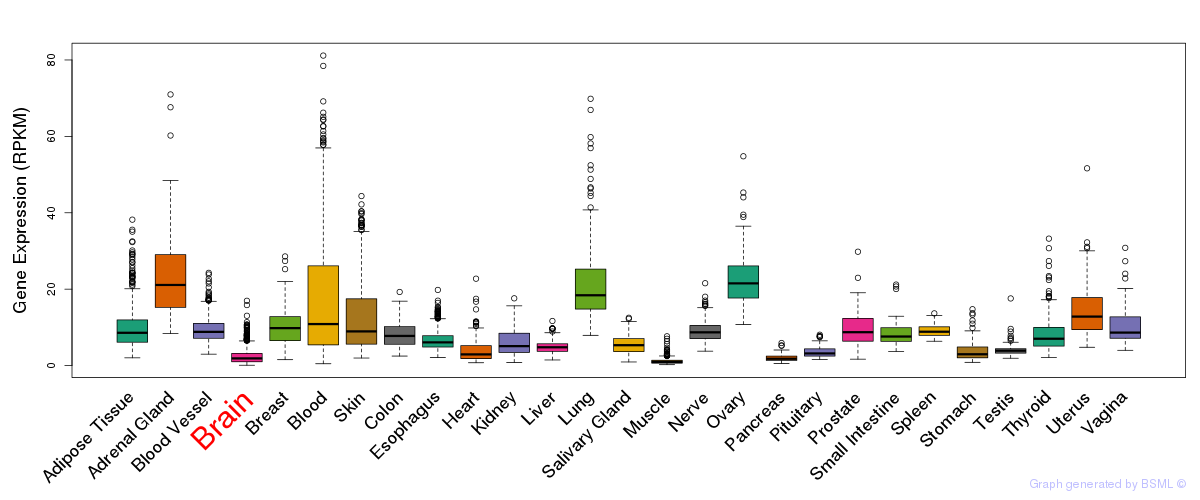
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FAM188B | 0.94 | 0.74 |
| GFAP | 0.83 | 0.89 |
| PLIN | 0.71 | 0.75 |
| RASL12 | 0.70 | 0.77 |
| ITGB4 | 0.67 | 0.79 |
| EFEMP1 | 0.67 | 0.79 |
| METTL7B | 0.65 | 0.68 |
| ACSM5 | 0.64 | 0.63 |
| NUPR1 | 0.64 | 0.77 |
| STK19P | 0.63 | 0.59 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| STMN1 | -0.36 | -0.63 |
| PLEKHO1 | -0.36 | -0.61 |
| ZBTB8A | -0.35 | -0.62 |
| NKIRAS2 | -0.35 | -0.62 |
| KIAA1949 | -0.35 | -0.58 |
| MPP3 | -0.34 | -0.61 |
| ZNF821 | -0.34 | -0.60 |
| TUBB2B | -0.34 | -0.58 |
| DUSP12 | -0.34 | -0.62 |
| BZW2 | -0.34 | -0.64 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004888 | transmembrane receptor activity | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0042802 | identical protein binding | IPI | 7536190 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006461 | protein complex assembly | TAS | 10875918 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0007165 | signal transduction | TAS | 9360929 | |
| GO:0008633 | activation of pro-apoptotic gene products | EXP | 14644197 | |
| GO:0006955 | immune response | IEA | - | |
| GO:0006916 | anti-apoptosis | TAS | 7510905 | |
| GO:0006915 | apoptosis | IEA | - | |
| GO:0008624 | induction of apoptosis by extracellular signals | EXP | 11048727 | |
| GO:0042981 | regulation of apoptosis | NAS | 7533181 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | NAS | 7533181 | |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005625 | soluble fraction | TAS | 7510905 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | EXP | 7530336 |7536190 |8521815 |8681376 |9721089 | |
| GO:0005886 | plasma membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARHGDIA | GDIA1 | MGC117248 | RHOGDI | RHOGDI-1 | Rho GDP dissociation inhibitor (GDI) alpha | Co-purification | BioGRID | 15659383 |
| BID | FP497 | MGC15319 | MGC42355 | BH3 interacting domain death agonist | Co-purification | BioGRID | 15659383 |
| BTK | AGMX1 | AT | ATK | BPK | IMD1 | MGC126261 | MGC126262 | PSCTK1 | XLA | Bruton agammaglobulinemia tyrosine kinase | - | HPRD | 9751072 |
| C14orf1 | ERG28 | NET51 | chromosome 14 open reading frame 1 | Two-hybrid | BioGRID | 16169070 |
| C1orf103 | FLJ11269 | RIF1 | RP11-96K19.1 | chromosome 1 open reading frame 103 | Two-hybrid | BioGRID | 16169070 |
| CASP10 | ALPS2 | FLICE2 | MCH4 | caspase 10, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 9045686 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | - | HPRD | 11101867 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | Affinity Capture-Western Co-purification | BioGRID | 9208847 |9325248 |15659383 |
| CASP8AP2 | CED-4 | FLASH | FLJ11208 | KIAA1315 | RIP25 | caspase 8 associated protein 2 | - | HPRD,BioGRID | 10235259 |
| CFLAR | CASH | CASP8AP1 | CLARP | Casper | FLAME | FLAME-1 | FLAME1 | FLIP | I-FLICE | MRIT | c-FLIP | c-FLIPL | c-FLIPR | c-FLIPS | CASP8 and FADD-like apoptosis regulator | Affinity Capture-Western | BioGRID | 9208847 |9325248 |
| CRMP1 | DPYSL1 | DRP-1 | DRP1 | collapsin response mediator protein 1 | Two-hybrid | BioGRID | 16169070 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | CTNNB1 (beta-catenin) interacts with the FAS promoter. | BIND | 15829968 |
| DAXX | BING2 | DAP6 | EAP1 | MGC126245 | MGC126246 | death-domain associated protein | Reconstituted Complex | BioGRID | 11483955 |
| DAXX | BING2 | DAP6 | EAP1 | MGC126245 | MGC126246 | death-domain associated protein | - | HPRD | 9215629 |11193028 |
| EEF1A1 | CCS-3 | CCS3 | EEF-1 | EEF1A | EF-Tu | EF1A | FLJ25721 | GRAF-1EF | HNGC:16303 | LENG7 | MGC102687 | MGC131894 | MGC16224 | PTI1 | eEF1A-1 | eukaryotic translation elongation factor 1 alpha 1 | Two-hybrid | BioGRID | 16169070 |
| EZR | CVIL | CVL | DKFZp762H157 | FLJ26216 | MGC1584 | VIL2 | ezrin | Co-purification | BioGRID | 15659383 |
| FADD | GIG3 | MGC8528 | MORT1 | Fas (TNFRSF6)-associated via death domain | Fas interacts with FADD. | BIND | 15383280 |
| FADD | GIG3 | MGC8528 | MORT1 | Fas (TNFRSF6)-associated via death domain | An unspecified isoform of Fas interacts with FADD. | BIND | 15665818 |
| FADD | GIG3 | MGC8528 | MORT1 | Fas (TNFRSF6)-associated via death domain | - | HPRD,BioGRID | 7538907 |8967952 |
| FADD | GIG3 | MGC8528 | MORT1 | Fas (TNFRSF6)-associated via death domain | - | HPRD | 7536190 |7538907 |8967952|7538907 |8967952 |
| FAF1 | CGI-03 | FLJ37524 | HFAF1s | UBXD12 | UBXN3A | hFAF1 | Fas (TNFRSF6) associated factor 1 | - | HPRD,BioGRID | 8524870 |9740801 |
| FAIM2 | KIAA0950 | LFG | NGP35 | NMP35 | TMBIM2 | Fas apoptotic inhibitory molecule 2 | - | HPRD,BioGRID | 10535980 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | Two-hybrid | BioGRID | 7538908 |
| FASLG | APT1LG1 | CD178 | CD95L | FASL | TNFSF6 | Fas ligand (TNF superfamily, member 6) | - | HPRD,BioGRID | 9126929 |9228058 |
| FBF1 | Alb | FBF-1 | FLJ00103 | Fas (TNFRSF6) binding factor 1 | - | HPRD,BioGRID | 10978533 |
| FEM1B | DKFZp451E0710 | FIAA | fem-1 homolog b (C. elegans) | - | HPRD | 10542291 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD,BioGRID | 8626376 |
| HIPK3 | DYRK6 | FIST3 | PKY | YAK1 | homeodomain interacting protein kinase 3 | - | HPRD,BioGRID | 11034606 |
| KAT5 | ESA1 | HTATIP | HTATIP1 | PLIP | TIP | TIP60 | cPLA2 | K(lysine) acetyltransferase 5 | HTATIP (Tip60) interacts with the FAS promoter. | BIND | 15829968 |
| LCK | YT16 | p56lck | pp58lck | lymphocyte-specific protein tyrosine kinase | - | HPRD,BioGRID | 8864141 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | Co-purification | BioGRID | 15659383 |
| MSN | - | moesin | Co-purification | BioGRID | 15659383 |
| NOL3 | ARC | CARD2 | MYC | MYP | NOP | NOP30 | nucleolar protein 3 (apoptosis repressor with CARD domain) | ARC interacts with an unspecified isoform of Fas. | BIND | 15383280 |
| NOL3 | ARC | CARD2 | MYC | MYP | NOP | NOP30 | nucleolar protein 3 (apoptosis repressor with CARD domain) | Phenotypic Suppression | BioGRID | 9560245 |
| PDCD6 | ALG-2 | FLJ46208 | MGC111017 | MGC119050 | MGC9123 | PEF1B | programmed cell death 6 | - | HPRD,BioGRID | 11606059 |
| PTPN13 | DKFZp686J1497 | FAP-1 | PNP1 | PTP-BAS | PTP-BL | PTP1E | PTPL1 | PTPLE | protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) | - | HPRD | 7536343 |9079683 |
| RHOA | ARH12 | ARHA | RHO12 | RHOH12 | ras homolog gene family, member A | Co-purification | BioGRID | 15659383 |
| RIPK1 | FLJ39204 | RIP | RIP1 | receptor (TNFRSF)-interacting serine-threonine kinase 1 | - | HPRD,BioGRID | 7538908 |9740801 |
| RUVBL2 | CGI-46 | ECP51 | INO80J | REPTIN | RVB2 | TIH2 | TIP48 | TIP49B | RuvB-like 2 (E. coli) | RUVBL2 (reptin) interacts with the FAS promoter. | BIND | 15829968 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | Reconstituted Complex Two-hybrid | BioGRID | 8906799 |11112409 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | Fas interacts with sentrin. | BIND | 11112409 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | - | HPRD | 8906799 |9740801 |
| TNFRSF10B | CD262 | DR5 | KILLER | KILLER/DR5 | TRAIL-R2 | TRAILR2 | TRICK2 | TRICK2A | TRICK2B | TRICKB | ZTNFR9 | tumor necrosis factor receptor superfamily, member 10b | Co-purification | BioGRID | 15659383 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | Co-purification | BioGRID | 15659383 |
| TNFSF12-TNFSF13 | TWE-PRIL | TNFSF12-TNFSF13 readthrough transcript | - | HPRD | 10706119 |
| TNFSF13 | APRIL | CD256 | TALL2 | TRDL-1 | UNQ383/PRO715 | ligand | tumor necrosis factor (ligand) superfamily, member 13 | Reconstituted Complex | BioGRID | 10706119 |
| TP63 | AIS | B(p51A) | B(p51B) | EEC3 | KET | LMS | NBP | OFC8 | RHS | SHFM4 | TP53CP | TP53L | TP73L | p40 | p51 | p53CP | p63 | p73H | p73L | tumor protein p63 | p63 interacts with CD95. | BIND | 15944736 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | - | HPRD | 8681376 |9427646 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | - | HPRD,BioGRID | 9257699 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG P53 SIGNALING PATHWAY | 69 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG NATURAL KILLER CELL MEDIATED CYTOTOXICITY | 137 | 92 | All SZGR 2.0 genes in this pathway |
| KEGG TYPE I DIABETES MELLITUS | 44 | 38 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG AUTOIMMUNE THYROID DISEASE | 53 | 49 | All SZGR 2.0 genes in this pathway |
| KEGG ALLOGRAFT REJECTION | 38 | 34 | All SZGR 2.0 genes in this pathway |
| KEGG GRAFT VERSUS HOST DISEASE | 42 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ASBCELL PATHWAY | 12 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CTL PATHWAY | 15 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FAS PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TCAPOPTOSIS PATHWAY | 11 | 8 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIVNEF PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL2RB PATHWAY | 38 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA KERATINOCYTE PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PML PATHWAY | 17 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HSP27 PATHWAY | 15 | 11 | All SZGR 2.0 genes in this pathway |
| SA CASPASE CASCADE | 19 | 13 | All SZGR 2.0 genes in this pathway |
| SA FAS SIGNALING | 9 | 5 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| PID P73PATHWAY | 79 | 59 | All SZGR 2.0 genes in this pathway |
| PID FAS PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID HIV NEF PATHWAY | 35 | 26 | All SZGR 2.0 genes in this pathway |
| PID TAP63 PATHWAY | 54 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | 13 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA UP | 294 | 178 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID UP | 25 | 18 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS DN | 186 | 114 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| HAHTOLA SEZARY SYNDROM UP | 98 | 58 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| LAU APOPTOSIS CDKN2A UP | 55 | 40 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS CDC25 DN | 51 | 35 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION UP | 211 | 136 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| BARRIER CANCER RELAPSE NORMAL SAMPLE UP | 32 | 21 | All SZGR 2.0 genes in this pathway |
| KERLEY RESPONSE TO CISPLATIN UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| MARKS HDAC TARGETS UP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| BAKER HEMATOPOIESIS STAT3 TARGETS | 16 | 12 | All SZGR 2.0 genes in this pathway |
| DUTTA APOPTOSIS VIA NFKB | 33 | 25 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 6 | 84 | 54 | All SZGR 2.0 genes in this pathway |
| SCHAVOLT TARGETS OF TP53 AND TP63 | 16 | 12 | All SZGR 2.0 genes in this pathway |
| DACOSTA LOW DOSE UV RESPONSE VIA ERCC3 XPCS UP | 18 | 10 | All SZGR 2.0 genes in this pathway |
| CEBALLOS TARGETS OF TP53 AND MYC DN | 38 | 31 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION | 149 | 101 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| ALCALA APOPTOSIS | 88 | 60 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A DN | 103 | 71 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| JEON SMAD6 TARGETS UP | 24 | 15 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 8 | 36 | 28 | All SZGR 2.0 genes in this pathway |
| DER IFN ALPHA RESPONSE UP | 74 | 48 | All SZGR 2.0 genes in this pathway |
| KANNAN TP53 TARGETS UP | 58 | 40 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| SASAKI ADULT T CELL LEUKEMIA | 176 | 122 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGLL VS IGLK UP | 42 | 24 | All SZGR 2.0 genes in this pathway |
| DER IFN GAMMA RESPONSE UP | 71 | 45 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| HALMOS CEBPA TARGETS UP | 52 | 34 | All SZGR 2.0 genes in this pathway |
| ABRAHAM ALPC VS MULTIPLE MYELOMA UP | 26 | 22 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| HOEGERKORP CD44 TARGETS TEMPORAL UP | 13 | 8 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| ONGUSAHA TP53 TARGETS | 38 | 23 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| CHUANG OXIDATIVE STRESS RESPONSE UP | 28 | 18 | All SZGR 2.0 genes in this pathway |
| WU HBX TARGETS 3 UP | 18 | 11 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| WU HBX TARGETS 1 UP | 16 | 10 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| YIH RESPONSE TO ARSENITE C1 | 24 | 14 | All SZGR 2.0 genes in this pathway |
| WU HBX TARGETS 2 UP | 23 | 13 | All SZGR 2.0 genes in this pathway |
| HAN JNK SINGALING DN | 39 | 27 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION DN | 281 | 179 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING DN | 36 | 24 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS UP | 43 | 28 | All SZGR 2.0 genes in this pathway |
| DE YY1 TARGETS DN | 92 | 64 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| SEKI INFLAMMATORY RESPONSE LPS UP | 77 | 56 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE D7 UP | 107 | 67 | All SZGR 2.0 genes in this pathway |
| SCHRAETS MLL TARGETS UP | 35 | 21 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| MCMURRAY TP53 HRAS COOPERATION RESPONSE DN | 67 | 46 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G123 DN | 51 | 30 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 UP | 176 | 110 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| HOELZEL NF1 TARGETS UP | 139 | 93 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS UP | 169 | 105 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 10HR UP | 54 | 38 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL MATURE DN | 99 | 74 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 2HR UP | 53 | 33 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR UP | 166 | 97 | All SZGR 2.0 genes in this pathway |
| GHANDHI DIRECT IRRADIATION UP | 110 | 68 | All SZGR 2.0 genes in this pathway |
| WARTERS RESPONSE TO IR SKIN | 83 | 44 | All SZGR 2.0 genes in this pathway |
| WARTERS IR RESPONSE 5GY | 47 | 23 | All SZGR 2.0 genes in this pathway |