Gene Page: IL3
Summary ?
| GeneID | 3562 |
| Symbol | IL3 |
| Synonyms | IL-3|MCGF|MULTI-CSF |
| Description | interleukin 3 |
| Reference | MIM:147740|HGNC:HGNC:6011|Ensembl:ENSG00000164399|HPRD:00987|Vega:OTTHUMG00000059640 |
| Gene type | protein-coding |
| Map location | 5q31.1 |
| Pascal p-value | 0.309 |
| Fetal beta | -0.109 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0032 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
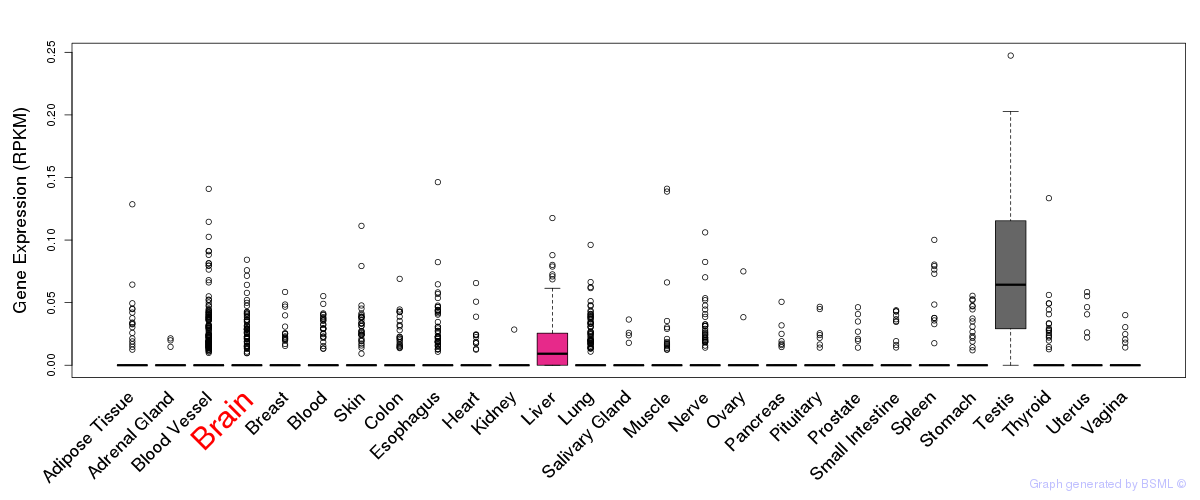
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| B3GAT1 | 0.71 | 0.69 |
| NUMB | 0.70 | 0.70 |
| MON1B | 0.69 | 0.72 |
| SPRY3 | 0.68 | 0.68 |
| BCR | 0.68 | 0.72 |
| PHACTR1 | 0.67 | 0.71 |
| CTNND2 | 0.67 | 0.69 |
| C6orf120 | 0.66 | 0.71 |
| RUNDC1 | 0.66 | 0.72 |
| NTRK3 | 0.66 | 0.72 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AL139819.3 | -0.49 | -0.57 |
| AF347015.21 | -0.47 | -0.51 |
| MT-CO2 | -0.44 | -0.46 |
| AF347015.31 | -0.44 | -0.46 |
| SYCP3 | -0.43 | -0.51 |
| GNG11 | -0.43 | -0.48 |
| HIGD1B | -0.42 | -0.43 |
| IL32 | -0.42 | -0.46 |
| AF347015.8 | -0.41 | -0.44 |
| AF347015.2 | -0.41 | -0.39 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005135 | interleukin-3 receptor binding | TAS | 2544122 | |
| GO:0005125 | cytokine activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 9736740 |
| GO:0007267 | cell-cell signaling | TAS | 3497400 | |
| GO:0008284 | positive regulation of cell proliferation | ISS | - | |
| GO:0006955 | immune response | IEA | - | |
| GO:0042523 | positive regulation of tyrosine phosphorylation of Stat5 protein | IDA | 9722506 | |
| GO:0043066 | negative regulation of apoptosis | IEA | - | |
| GO:0019221 | cytokine-mediated signaling pathway | IEA | - | |
| GO:0045740 | positive regulation of DNA replication | IDA | 9722506 | |
| GO:0045885 | positive regulation of survival gene product expression | ISS | - | |
| GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | ISS | - | |
| GO:0046330 | positive regulation of JNK cascade | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | ISS | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY | 155 | 105 | All SZGR 2.0 genes in this pathway |
| KEGG HEMATOPOIETIC CELL LINEAGE | 88 | 60 | All SZGR 2.0 genes in this pathway |
| KEGG FC EPSILON RI SIGNALING PATHWAY | 79 | 58 | All SZGR 2.0 genes in this pathway |
| KEGG ASTHMA | 30 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CYTOKINE PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INFLAM PATHWAY | 29 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DC PATHWAY | 22 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ERYTH PATHWAY | 15 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL17 PATHWAY | 17 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL3 PATHWAY | 15 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BAD PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| BIOCARTA STEM PATHWAY | 15 | 13 | All SZGR 2.0 genes in this pathway |
| PID NFAT TFPATHWAY | 47 | 39 | All SZGR 2.0 genes in this pathway |
| PID IL3 PATHWAY | 27 | 19 | All SZGR 2.0 genes in this pathway |
| PID TCR CALCIUM PATHWAY | 29 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ILS | 107 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME IL 3 5 AND GM CSF SIGNALING | 43 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME IL RECEPTOR SHC SIGNALING | 27 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME IL 2 SIGNALING | 41 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| DEBOSSCHER NFKB TARGETS REPRESSED BY GLUCOCORTICOIDS | 24 | 18 | All SZGR 2.0 genes in this pathway |
| HOFMANN CELL LYMPHOMA UP | 50 | 35 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND TBH | 60 | 42 | All SZGR 2.0 genes in this pathway |
| MCCLUNG COCAIN REWARD 4WK | 75 | 47 | All SZGR 2.0 genes in this pathway |
| RAY ALZHEIMERS DISEASE | 13 | 7 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR DN | 88 | 59 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |