| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION
| 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY
| 155 | 105 | All SZGR 2.0 genes in this pathway |
| KEGG HEMATOPOIETIC CELL LINEAGE
| 88 | 60 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY
| 108 | 89 | All SZGR 2.0 genes in this pathway |
| KEGG FC EPSILON RI SIGNALING PATHWAY
| 79 | 58 | All SZGR 2.0 genes in this pathway |
| KEGG INTESTINAL IMMUNE NETWORK FOR IGA PRODUCTION
| 48 | 36 | All SZGR 2.0 genes in this pathway |
| KEGG LEISHMANIA INFECTION
| 72 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG ASTHMA
| 30 | 25 | All SZGR 2.0 genes in this pathway |
| KEGG AUTOIMMUNE THYROID DISEASE
| 53 | 49 | All SZGR 2.0 genes in this pathway |
| KEGG ALLOGRAFT REJECTION
| 38 | 34 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ASBCELL PATHWAY
| 12 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CYTOKINE PATHWAY
| 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INFLAM PATHWAY
| 29 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DC PATHWAY
| 22 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GATA3 PATHWAY
| 16 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL4 PATHWAY
| 11 | 8 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL5 PATHWAY
| 10 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA STEM PATHWAY
| 15 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TOB1 PATHWAY
| 21 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NKT PATHWAY
| 29 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TH1TH2 PATHWAY
| 19 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA 41BB PATHWAY
| 17 | 15 | All SZGR 2.0 genes in this pathway |
| ST INTERLEUKIN 4 PATHWAY
| 26 | 18 | All SZGR 2.0 genes in this pathway |
| PID NOTCH PATHWAY
| 59 | 49 | All SZGR 2.0 genes in this pathway |
| PID IL4 2PATHWAY
| 65 | 43 | All SZGR 2.0 genes in this pathway |
| PID CD40 PATHWAY
| 31 | 26 | All SZGR 2.0 genes in this pathway |
| PID IL12 2PATHWAY
| 63 | 54 | All SZGR 2.0 genes in this pathway |
| PID NFAT TFPATHWAY
| 47 | 39 | All SZGR 2.0 genes in this pathway |
| PID REG GR PATHWAY
| 82 | 60 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY
| 70 | 60 | All SZGR 2.0 genes in this pathway |
| PID IL2 STAT5 PATHWAY
| 30 | 23 | All SZGR 2.0 genes in this pathway |
| PID TCR CALCIUM PATHWAY
| 29 | 23 | All SZGR 2.0 genes in this pathway |
| ROYLANCE BREAST CANCER 16Q COPY NUMBER UP
| 63 | 44 | All SZGR 2.0 genes in this pathway |
| DEBOSSCHER NFKB TARGETS REPRESSED BY GLUCOCORTICOIDS
| 24 | 18 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK
| 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK
| 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK
| 779 | 480 | All SZGR 2.0 genes in this pathway |
| BRUNO HEMATOPOIESIS
| 66 | 48 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION
| 405 | 264 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM
| 302 | 191 | All SZGR 2.0 genes in this pathway |
| JU AGING TERC TARGETS UP
| 10 | 6 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE UP
| 163 | 102 | All SZGR 2.0 genes in this pathway |
| BUDHU LIVER CANCER METASTASIS UP
| 10 | 7 | All SZGR 2.0 genes in this pathway |
| ONO AML1 TARGETS DN
| 41 | 25 | All SZGR 2.0 genes in this pathway |
| ONO FOXP3 TARGETS DN
| 42 | 23 | All SZGR 2.0 genes in this pathway |
| WORSCHECH TUMOR EVASION AND TOLEROGENICITY UP
| 30 | 19 | All SZGR 2.0 genes in this pathway |
| GREGORY SYNTHETIC LETHAL WITH IMATINIB
| 145 | 83 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A
| 898 | 516 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY
| 1725 | 838 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS
| 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED
| 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME
| 1028 | 559 | All SZGR 2.0 genes in this pathway |
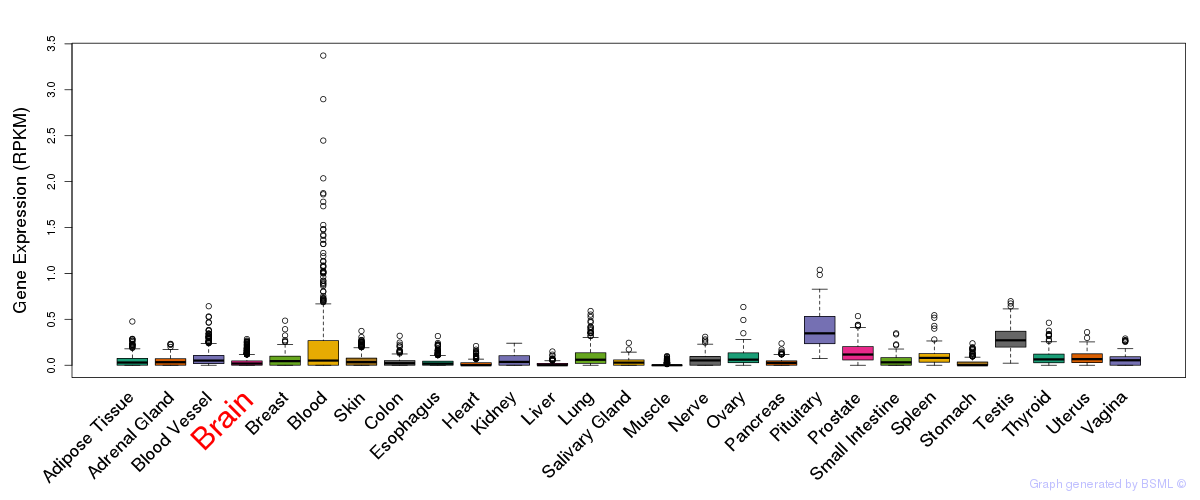
 eQTL annotation
eQTL annotation