Gene Page: LHX1
Summary ?
| GeneID | 3975 |
| Symbol | LHX1 |
| Synonyms | LIM-1|LIM1 |
| Description | LIM homeobox 1 |
| Reference | MIM:601999|HGNC:HGNC:6593|Ensembl:ENSG00000273706|HPRD:09067|Vega:OTTHUMG00000188456 |
| Gene type | protein-coding |
| Map location | 17q12 |
| Pascal p-value | 0.18 |
| Fetal beta | 0.635 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6687849 | chr1 | 175904808 | LHX1 | 3975 | 0.18 | trans | ||
| rs2502827 | chr1 | 176044216 | LHX1 | 3975 | 0.02 | trans | ||
| rs17572651 | chr1 | 218943612 | LHX1 | 3975 | 1.642E-4 | trans | ||
| rs6734526 | chr2 | 5616370 | LHX1 | 3975 | 0.02 | trans | ||
| rs6753473 | chr2 | 26526418 | LHX1 | 3975 | 0.12 | trans | ||
| rs17028363 | chr2 | 41913161 | LHX1 | 3975 | 0.18 | trans | ||
| rs16829545 | chr2 | 151977407 | LHX1 | 3975 | 6.31E-36 | trans | ||
| rs16841750 | chr2 | 158288461 | LHX1 | 3975 | 2.792E-4 | trans | ||
| rs3845734 | chr2 | 171125572 | LHX1 | 3975 | 0.02 | trans | ||
| rs7584986 | chr2 | 184111432 | LHX1 | 3975 | 2.139E-11 | trans | ||
| rs6741060 | chr2 | 217169088 | LHX1 | 3975 | 0 | trans | ||
| rs9810143 | chr3 | 5060209 | LHX1 | 3975 | 3.73E-4 | trans | ||
| rs6797307 | chr3 | 8601563 | LHX1 | 3975 | 0.03 | trans | ||
| rs7692715 | chr4 | 125335209 | LHX1 | 3975 | 0.15 | trans | ||
| rs2183142 | chr4 | 159232695 | LHX1 | 3975 | 0.03 | trans | ||
| rs170776 | chr4 | 173276735 | LHX1 | 3975 | 0.02 | trans | ||
| rs1396222 | chr4 | 173279496 | LHX1 | 3975 | 0.01 | trans | ||
| rs335993 | chr4 | 173321789 | LHX1 | 3975 | 0.03 | trans | ||
| rs335980 | chr4 | 173329784 | LHX1 | 3975 | 0.01 | trans | ||
| rs335982 | chr4 | 173330945 | LHX1 | 3975 | 0.03 | trans | ||
| rs336016 | chr4 | 173366576 | LHX1 | 3975 | 0.14 | trans | ||
| rs337984 | chr4 | 173411662 | LHX1 | 3975 | 0.03 | trans | ||
| rs17762315 | chr5 | 76807576 | LHX1 | 3975 | 0.02 | trans | ||
| rs10491487 | chr5 | 80323367 | LHX1 | 3975 | 0.03 | trans | ||
| rs10474151 | chr5 | 84286402 | LHX1 | 3975 | 0.02 | trans | ||
| rs13360496 | 0 | LHX1 | 3975 | 0.11 | trans | |||
| rs13191953 | chr6 | 24151042 | LHX1 | 3975 | 0.04 | trans | ||
| rs12196880 | chr6 | 24313746 | LHX1 | 3975 | 0.04 | trans | ||
| rs10806988 | chr6 | 24341768 | LHX1 | 3975 | 0.11 | trans | ||
| rs16890367 | chr6 | 38078448 | LHX1 | 3975 | 0.01 | trans | ||
| rs12706918 | chr7 | 80266147 | LHX1 | 3975 | 0.16 | trans | ||
| rs7787830 | chr7 | 98797019 | LHX1 | 3975 | 0.18 | trans | ||
| rs6996695 | chr8 | 77540580 | LHX1 | 3975 | 0.01 | trans | ||
| rs3118341 | chr9 | 25185518 | LHX1 | 3975 | 0.02 | trans | ||
| rs9406868 | chr9 | 25223372 | LHX1 | 3975 | 0.03 | trans | ||
| rs11139334 | chr9 | 84209393 | LHX1 | 3975 | 0 | trans | ||
| rs2393316 | chr10 | 59333070 | LHX1 | 3975 | 0.01 | trans | ||
| rs11061703 | chr12 | 1436977 | LHX1 | 3975 | 0.07 | trans | ||
| rs7337967 | chr13 | 53519044 | LHX1 | 3975 | 0.1 | trans | ||
| rs16955618 | chr15 | 29937543 | LHX1 | 3975 | 1.572E-45 | trans | ||
| rs2077735 | chr15 | 58479024 | LHX1 | 3975 | 0.08 | trans | ||
| rs11873184 | chr18 | 1584081 | LHX1 | 3975 | 0.01 | trans | ||
| rs12458173 | chr18 | 31430166 | LHX1 | 3975 | 0.07 | trans | ||
| rs17681878 | chr18 | 34767223 | LHX1 | 3975 | 0.05 | trans | ||
| rs749043 | chr20 | 46995700 | LHX1 | 3975 | 0.19 | trans | ||
| rs1041786 | chr21 | 22617710 | LHX1 | 3975 | 1.039E-5 | trans | ||
| rs16986332 | chrX | 10958354 | LHX1 | 3975 | 0.13 | trans | ||
| rs16990575 | chrX | 32691327 | LHX1 | 3975 | 0.03 | trans |
Section II. Transcriptome annotation
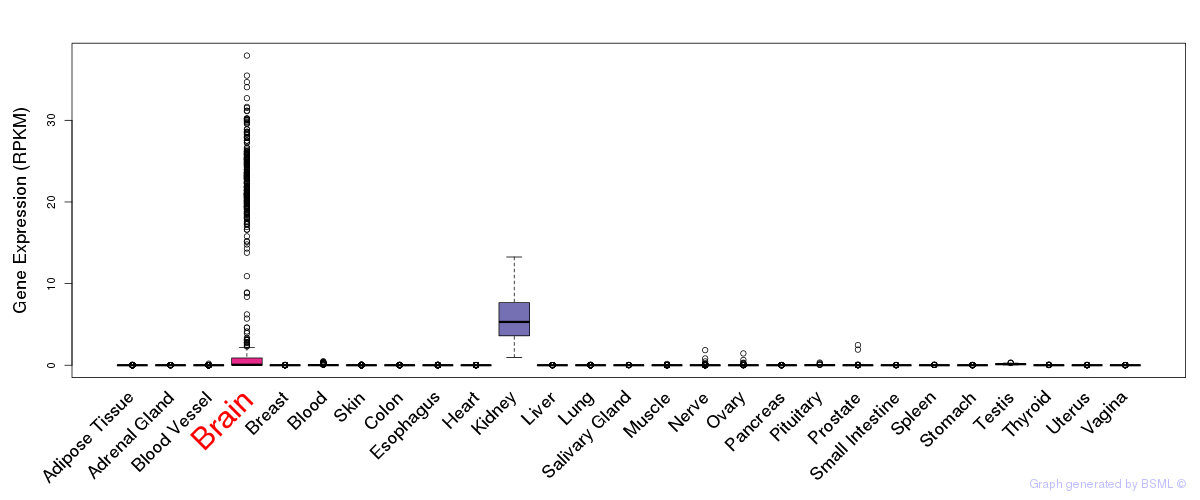
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ARAP1 | 0.83 | 0.83 |
| LTBP3 | 0.80 | 0.81 |
| LZTS2 | 0.77 | 0.77 |
| PLXNB1 | 0.76 | 0.75 |
| RNPEPL1 | 0.76 | 0.75 |
| DNM2 | 0.75 | 0.72 |
| SLC38A10 | 0.75 | 0.72 |
| TLN1 | 0.74 | 0.74 |
| PLOD3 | 0.73 | 0.76 |
| TMEM104 | 0.73 | 0.71 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C18orf32 | -0.39 | -0.47 |
| LY96 | -0.38 | -0.26 |
| GLMN | -0.37 | -0.39 |
| CLEC2B | -0.37 | -0.19 |
| SYCP3 | -0.37 | -0.25 |
| NOX1 | -0.37 | -0.29 |
| AC087071.1 | -0.37 | -0.21 |
| CAPZA2 | -0.36 | -0.40 |
| TMEM126A | -0.36 | -0.38 |
| AL050337.1 | -0.36 | -0.25 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | NAS | 9212161 | |
| GO:0003702 | RNA polymerase II transcription factor activity | TAS | 9212161 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0021702 | cerebellar Purkinje cell differentiation | IEA | neuron, GABA, Brain (GO term level: 15) | - |
| GO:0021549 | cerebellum development | IEA | Brain (GO term level: 10) | - |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 9212161 |
| GO:0001706 | endoderm formation | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0009887 | organ morphogenesis | TAS | 7700351 | |
| GO:0009791 | post-embryonic development | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0021937 | Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| TANAKA METHYLATED IN ESOPHAGEAL CARCINOMA | 103 | 58 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 2 DN | 51 | 42 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN | 200 | 112 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB TARGETS | 74 | 41 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT DN | 185 | 111 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 2 | 50 | 34 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| LEIN CEREBELLUM MARKERS | 85 | 47 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| MATZUK IMPLANTATION AND UTERINE | 22 | 15 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3 | 269 | 159 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |