Gene Page: LNPEP
Summary ?
| GeneID | 4012 |
| Symbol | LNPEP |
| Synonyms | CAP|IRAP|P-LAP|PLAP |
| Description | leucyl/cystinyl aminopeptidase |
| Reference | MIM:151300|HGNC:HGNC:6656|Ensembl:ENSG00000113441|HPRD:01041|HPRD:08255|Vega:OTTHUMG00000128719 |
| Gene type | protein-coding |
| Map location | 5q15 |
| Pascal p-value | 0.005 |
| DEG p-value | DEG:Sanders_2014:DS1_p=0.154:DS1_beta=0.020700:DS2_p=2.64e-01:DS2_beta=0.053:DS2_FDR=5.19e-01 |
| Fetal beta | -0.191 |
| DMG | 2 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Sanders_2013 | Microarray | Whole-genome gene expression profiles using microarrays on lymphoblastoid cell lines (LCLs) from 413 cases and 446 controls. | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03635536 | 5 | 96271340 | LNPEP | 2.039E-4 | -0.305 | 0.035 | DMG:Wockner_2014 |
| cg16189644 | 5 | 96271684 | LNPEP | 4.64E-9 | -0.014 | 2.7E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs149481 | chr5 | 96114345 | LNPEP | 4012 | 0.15 | cis | ||
| rs1230358 | chr5 | 96211740 | LNPEP | 4012 | 0 | cis | ||
| rs2549779 | chr5 | 96216386 | LNPEP | 4012 | 0.01 | cis | ||
| rs2549780 | chr5 | 96216783 | LNPEP | 4012 | 0.02 | cis | ||
| rs11135482 | chr5 | 96221425 | LNPEP | 4012 | 7.933E-5 | cis | ||
| rs2278019 | chr5 | 96225251 | LNPEP | 4012 | 0 | cis | ||
| rs10434709 | chr5 | 96225773 | LNPEP | 4012 | 9.233E-5 | cis | ||
| rs2548533 | chr5 | 96238400 | LNPEP | 4012 | 1.462E-5 | cis | ||
| rs2549794 | chr5 | 96244548 | LNPEP | 4012 | 0 | cis | ||
| rs7736466 | chr5 | 96289710 | LNPEP | 4012 | 6.621E-6 | cis | ||
| rs3909451 | chr5 | 96295120 | LNPEP | 4012 | 3.871E-4 | cis | ||
| rs27307 | chr5 | 96338504 | LNPEP | 4012 | 2.507E-4 | cis | ||
| rs27290 | chr5 | 96350087 | LNPEP | 4012 | 3.17E-4 | cis | ||
| rs27993 | chr5 | 96355602 | LNPEP | 4012 | 2.467E-4 | cis | ||
| rs27300 | chr5 | 96363406 | LNPEP | 4012 | 4.582E-4 | cis | ||
| rs1230358 | chr5 | 96211740 | LNPEP | 4012 | 0.12 | trans | ||
| rs11135482 | chr5 | 96221425 | LNPEP | 4012 | 0.01 | trans | ||
| rs2278019 | chr5 | 96225251 | LNPEP | 4012 | 0.2 | trans | ||
| rs10434709 | chr5 | 96225773 | LNPEP | 4012 | 0.01 | trans | ||
| rs2548533 | chr5 | 96238400 | LNPEP | 4012 | 0 | trans | ||
| rs2549794 | chr5 | 96244548 | LNPEP | 4012 | 0.09 | trans | ||
| rs7736466 | chr5 | 96289710 | LNPEP | 4012 | 0 | trans | ||
| rs3909451 | chr5 | 96295120 | LNPEP | 4012 | 0.03 | trans | ||
| rs27307 | chr5 | 96338504 | LNPEP | 4012 | 0.02 | trans | ||
| rs27290 | chr5 | 96350087 | LNPEP | 4012 | 0.03 | trans | ||
| rs27993 | chr5 | 96355602 | LNPEP | 4012 | 0.02 | trans | ||
| rs27300 | chr5 | 96363406 | LNPEP | 4012 | 0.04 | trans |
Section II. Transcriptome annotation
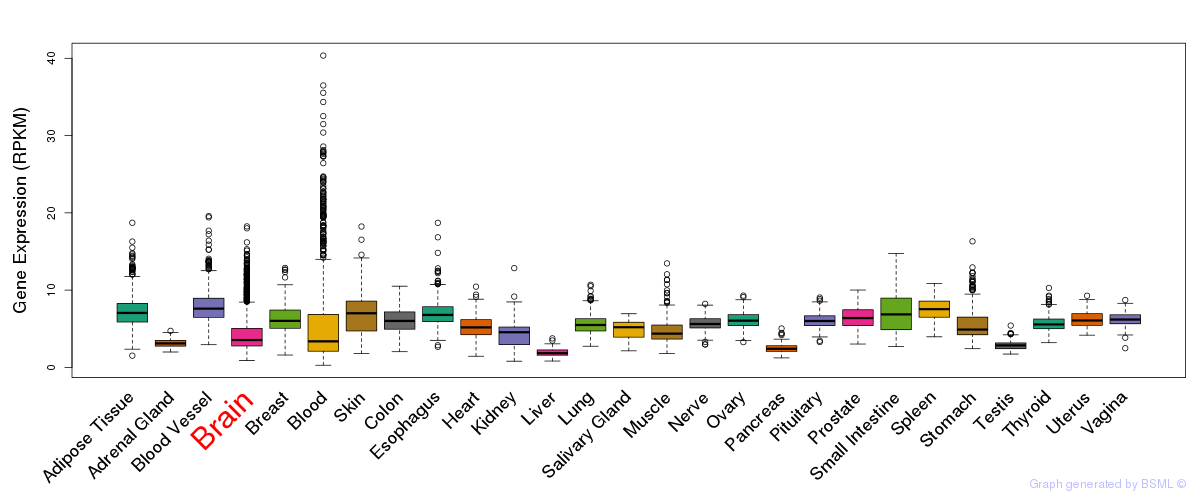
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C1QTNF4 | 0.86 | 0.87 |
| YDJC | 0.86 | 0.90 |
| HSPBP1 | 0.85 | 0.84 |
| SSBP4 | 0.84 | 0.88 |
| SLC39A3 | 0.83 | 0.83 |
| ZNF580 | 0.82 | 0.87 |
| GP1BB | 0.82 | 0.84 |
| FBXW5 | 0.80 | 0.83 |
| B4GALT2 | 0.80 | 0.77 |
| NUDT16L1 | 0.79 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NSBP1 | -0.45 | -0.52 |
| AL139819.3 | -0.45 | -0.48 |
| AF347015.8 | -0.43 | -0.36 |
| AF347015.18 | -0.43 | -0.38 |
| MT-ATP8 | -0.42 | -0.39 |
| AF347015.21 | -0.41 | -0.34 |
| AF347015.26 | -0.41 | -0.35 |
| EIF5B | -0.41 | -0.49 |
| AF347015.33 | -0.40 | -0.35 |
| AF347015.2 | -0.40 | -0.32 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG RENIN ANGIOTENSIN SYSTEM | 17 | 10 | All SZGR 2.0 genes in this pathway |
| SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | 51 | 41 | All SZGR 2.0 genes in this pathway |
| PID INSULIN GLUCOSE PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | 76 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME ENDOSOMAL VACUOLAR PATHWAY | 9 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | 212 | 129 | All SZGR 2.0 genes in this pathway |
| LIU CMYB TARGETS UP | 165 | 106 | All SZGR 2.0 genes in this pathway |
| LIU VMYB TARGETS UP | 127 | 78 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM UP | 176 | 111 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A DN | 110 | 78 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE DN | 123 | 76 | All SZGR 2.0 genes in this pathway |
| BILD SRC ONCOGENIC SIGNATURE | 62 | 38 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| LIN NPAS4 TARGETS UP | 163 | 100 | All SZGR 2.0 genes in this pathway |
| LEE DIFFERENTIATING T LYMPHOCYTE | 200 | 115 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| VISALA AGING LYMPHOCYTE UP | 10 | 7 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G1 S | 147 | 76 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 1 DN | 63 | 39 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |