Gene Page: ARL2
Summary ?
| GeneID | 402 |
| Symbol | ARL2 |
| Synonyms | ARFL2 |
| Description | ADP ribosylation factor like GTPase 2 |
| Reference | MIM:601175|HGNC:HGNC:693|Ensembl:ENSG00000213465|HPRD:11869|Vega:OTTHUMG00000165728 |
| Gene type | protein-coding |
| Map location | 11q13 |
| Sherlock p-value | 0.293 |
| Fetal beta | -1.201 |
| DMG | 2 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg21965516 | 11 | 64781475 | ARL2 | 1.184E-4 | -0.373 | 0.029 | DMG:Wockner_2014 |
| cg19017420 | 11 | 64781299 | ARL2 | -0.019 | 0.91 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
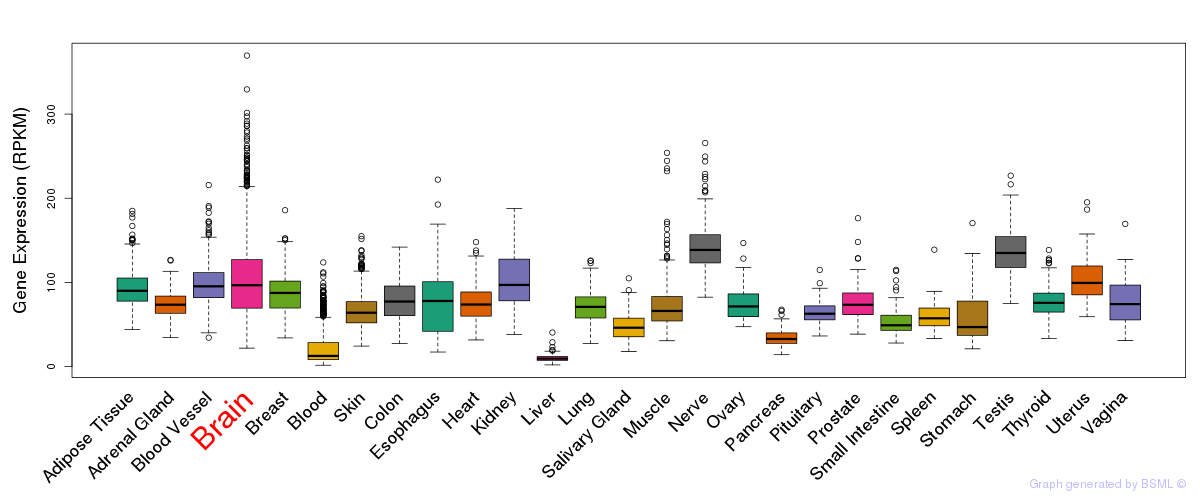
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TUBGCP3 | 0.91 | 0.90 |
| PHF8 | 0.89 | 0.90 |
| GPATCH8 | 0.88 | 0.89 |
| FKBP15 | 0.88 | 0.87 |
| NUP98 | 0.88 | 0.89 |
| SUPT6H | 0.88 | 0.89 |
| UBE4B | 0.88 | 0.88 |
| GIT2 | 0.88 | 0.89 |
| ARIH2 | 0.88 | 0.88 |
| ADD1 | 0.87 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.73 | -0.79 |
| MT-CO2 | -0.71 | -0.74 |
| AF347015.31 | -0.71 | -0.74 |
| IFI27 | -0.71 | -0.72 |
| HIGD1B | -0.70 | -0.73 |
| FXYD1 | -0.70 | -0.72 |
| C1orf54 | -0.69 | -0.79 |
| VAMP5 | -0.68 | -0.71 |
| CXCL14 | -0.66 | -0.69 |
| MYL3 | -0.65 | -0.67 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARL2BP | BART1 | ADP-ribosylation factor-like 2 binding protein | - | HPRD,BioGRID | 11809823 |
| PDE6D | PDED | phosphodiesterase 6D, cGMP-specific, rod, delta | - | HPRD,BioGRID | 11980706 |
| PPP2CA | PP2Ac | PP2CA | RP-C | protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform | Co-purification | BioGRID | 12912990 |
| PPP2CB | PP2CB | protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform | Co-purification | BioGRID | 12912990 |
| PPP2R1B | MGC26454 | PR65B | protein phosphatase 2 (formerly 2A), regulatory subunit A, beta isoform | - | HPRD | 12912990 |
| PPP2R2B | B55-BETA | FLJ95686 | MGC24888 | PP2A-B55BETA | PP2A-PR55B | PP2AB-BETA | PP2APR55-BETA | PR2AB-BETA | PR2AB55-BETA | PR2APR55-BETA | PR52B | PR55-BETA | SCA12 | protein phosphatase 2 (formerly 2A), regulatory subunit B, beta isoform | - | HPRD | 12912990 |
| PPP2R2C | B55-GAMMA | IMYPNO | IMYPNO1 | MGC33570 | PR52 | PR55G | protein phosphatase 2 (formerly 2A), regulatory subunit B, gamma isoform | - | HPRD,BioGRID | 12912990 |
| PPP2R5D | B56D | MGC2134 | MGC8949 | protein phosphatase 2, regulatory subunit B', delta isoform | Co-purification | BioGRID | 12912990 |
| SLC25A4 | AAC1 | ANT | ANT1 | PEO2 | PEO3 | T1 | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 | - | HPRD | 11809823 |
| TBCD | KIAA0988 | tubulin folding cofactor D | - | HPRD,BioGRID | 10831612 |12912990 |
| UNC119 | HRG4 | unc-119 homolog (C. elegans) | HRG4 interacts with ARL2. | BIND | 12527357 |
| UNC119 | HRG4 | unc-119 homolog (C. elegans) | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12527357 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER UP | 142 | 96 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW DN | 165 | 107 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH DN | 180 | 110 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA UP | 146 | 104 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION DN | 184 | 132 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| BONCI TARGETS OF MIR15A AND MIR16 1 | 91 | 75 | All SZGR 2.0 genes in this pathway |
| HOFFMANN IMMATURE TO MATURE B LYMPHOCYTE UP | 43 | 34 | All SZGR 2.0 genes in this pathway |