Gene Page: LTA
Summary ?
| GeneID | 4049 |
| Symbol | LTA |
| Synonyms | LT|TNFB|TNFSF1|TNLG1E |
| Description | lymphotoxin alpha |
| Reference | MIM:153440|HGNC:HGNC:6709|Ensembl:ENSG00000226979|HPRD:01084|Vega:OTTHUMG00000031135 |
| Gene type | protein-coding |
| Map location | 6p21.3 |
| Pascal p-value | 9.851E-8 |
| Fetal beta | 0.006 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
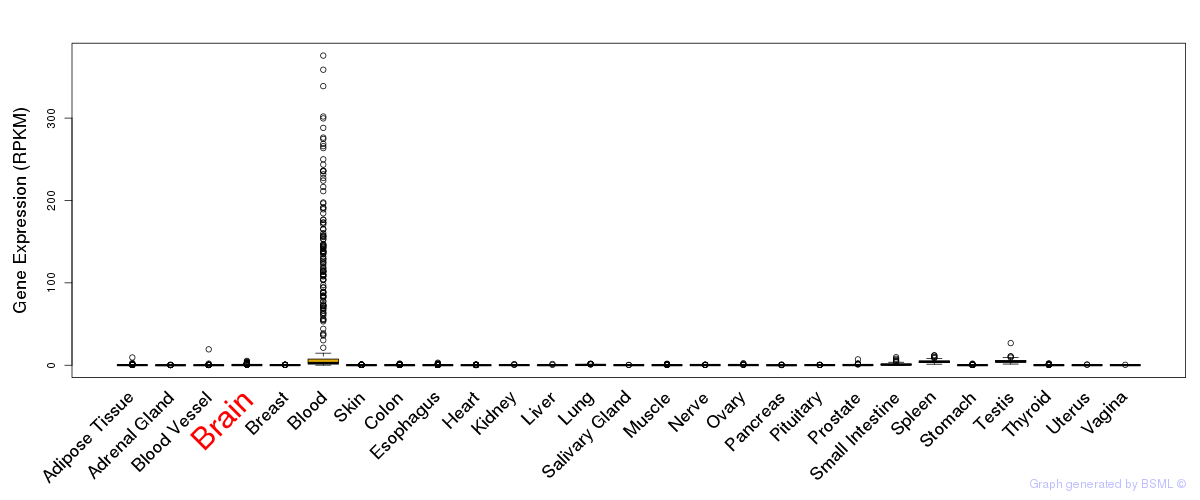
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| WDR42A | 0.89 | 0.84 |
| APLP2 | 0.88 | 0.86 |
| APP | 0.86 | 0.85 |
| SRPR | 0.85 | 0.86 |
| AARS | 0.85 | 0.84 |
| TK2 | 0.85 | 0.83 |
| HDHD2 | 0.85 | 0.81 |
| PROSC | 0.85 | 0.83 |
| RAB5B | 0.84 | 0.81 |
| SYT11 | 0.84 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.53 | -0.27 |
| IL32 | -0.50 | -0.33 |
| AF347015.18 | -0.49 | -0.25 |
| AC098691.2 | -0.48 | -0.39 |
| FAM159B | -0.47 | -0.46 |
| GNG11 | -0.44 | -0.29 |
| MT-CO2 | -0.44 | -0.26 |
| C1orf54 | -0.43 | -0.30 |
| AF347015.2 | -0.42 | -0.22 |
| C1orf61 | -0.42 | -0.37 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005125 | cytokine activity | IEA | - | |
| GO:0005164 | tumor necrosis factor receptor binding | IEA | - | |
| GO:0004197 | cysteine-type endopeptidase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006508 | proteolysis | IEA | - | |
| GO:0007267 | cell-cell signaling | TAS | 6334807 | |
| GO:0006917 | induction of apoptosis | TAS | 6334807 | |
| GO:0007165 | signal transduction | TAS | 6334807 | |
| GO:0006959 | humoral immune response | IEA | - | |
| GO:0006955 | immune response | IEA | - | |
| GO:0048535 | lymph node development | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | IEA | - | |
| GO:0016020 | membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG ANTIGEN PROCESSING AND PRESENTATION | 89 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG TYPE I DIABETES MELLITUS | 44 | 38 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CYTOKINE PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INFLAM PATHWAY | 29 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA STRESS PATHWAY | 25 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TNFR2 PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| PID IL4 2PATHWAY | 65 | 43 | All SZGR 2.0 genes in this pathway |
| PID IL2 STAT5 PATHWAY | 30 | 23 | All SZGR 2.0 genes in this pathway |
| WANG BARRETTS ESOPHAGUS AND ESOPHAGUS CANCER UP | 26 | 12 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE EARLY | 66 | 48 | All SZGR 2.0 genes in this pathway |
| FUNG IL2 SIGNALING 1 | 11 | 7 | All SZGR 2.0 genes in this pathway |
| FUNG IL2 TARGETS WITH STAT5 BINDING SITES | 7 | 5 | All SZGR 2.0 genes in this pathway |
| YAN ESCAPE FROM ANOIKIS | 24 | 19 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING VIA SMAD4 DN | 66 | 38 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| SHIPP DLBCL CURED VS FATAL UP | 39 | 22 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGLL VS IGLK DN | 24 | 18 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP | 203 | 130 | All SZGR 2.0 genes in this pathway |
| XU CREBBP TARGETS DN | 44 | 31 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING UP | 101 | 76 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION MUSCLE UP | 43 | 33 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| VANASSE BCL2 TARGETS DN | 74 | 50 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF ICP WITH H3K27ME3 | 206 | 108 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS ICP WITH H3K4ME3 AND H327ME3 | 126 | 83 | All SZGR 2.0 genes in this pathway |
| KUROZUMI RESPONSE TO ONCOCYTIC VIRUS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| KUROZUMI RESPONSE TO ONCOCYTIC VIRUS AND CYCLIC RGD | 21 | 15 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 AND H3K27ME3 | 137 | 85 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR DN | 88 | 59 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP | 194 | 133 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR DN | 114 | 69 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |