Gene Page: LY9
Summary ?
| GeneID | 4063 |
| Symbol | LY9 |
| Synonyms | CD229|SLAMF3|hly9|mLY9 |
| Description | lymphocyte antigen 9 |
| Reference | MIM:600684|HGNC:HGNC:6730|Ensembl:ENSG00000122224|HPRD:02817|Vega:OTTHUMG00000024007 |
| Gene type | protein-coding |
| Map location | 1q23.3 |
| Fetal beta | -0.46 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
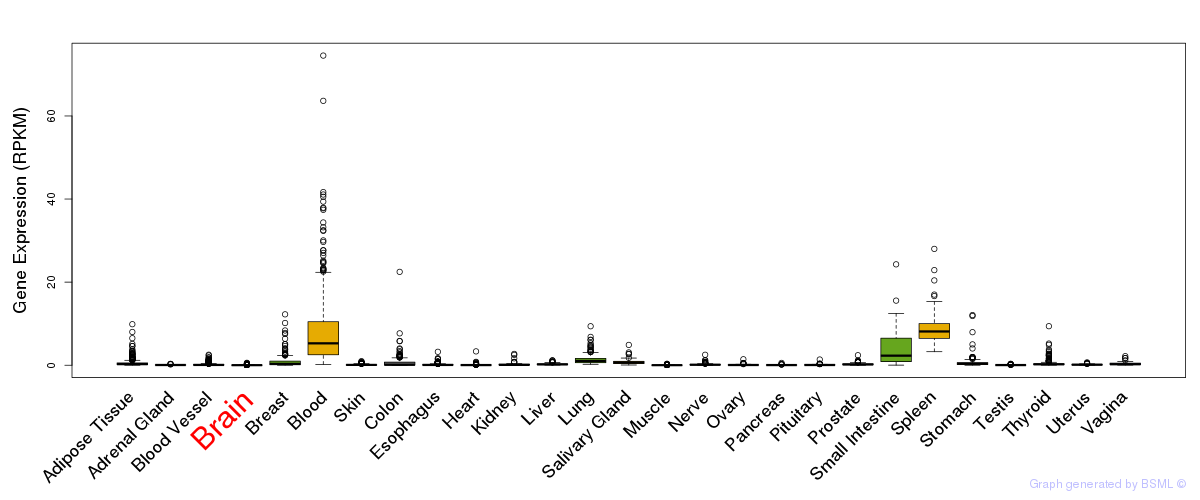
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| STIM1 | 0.91 | 0.90 |
| MAST3 | 0.91 | 0.86 |
| ACTN1 | 0.90 | 0.84 |
| IQSEC1 | 0.90 | 0.88 |
| PACSIN1 | 0.89 | 0.82 |
| SIRPA | 0.89 | 0.88 |
| CACNB2 | 0.89 | 0.81 |
| SYNPO | 0.88 | 0.81 |
| C2orf55 | 0.88 | 0.79 |
| KCNMA1 | 0.88 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.51 | -0.62 |
| C9orf46 | -0.50 | -0.57 |
| HEBP2 | -0.49 | -0.66 |
| RPL23A | -0.48 | -0.59 |
| RPS8 | -0.47 | -0.60 |
| RPS18 | -0.47 | -0.57 |
| C21orf57 | -0.47 | -0.53 |
| DYNLT1 | -0.47 | -0.62 |
| TRAF4 | -0.47 | -0.54 |
| RPL32 | -0.47 | -0.56 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003674 | molecular_function | ND | - | |
| GO:0005515 | protein binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007155 | cell adhesion | IEA | - | |
| GO:0016064 | immunoglobulin mediated immune response | NAS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA DN | 136 | 86 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 UP | 201 | 125 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL DN | 226 | 132 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| SHIPP DLBCL VS FOLLICULAR LYMPHOMA DN | 45 | 28 | All SZGR 2.0 genes in this pathway |
| LEE AGING NEOCORTEX DN | 80 | 49 | All SZGR 2.0 genes in this pathway |
| LIAN LIPA TARGETS 6M | 74 | 47 | All SZGR 2.0 genes in this pathway |
| LIAN LIPA TARGETS 3M | 59 | 36 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| KONDO EZH2 TARGETS | 245 | 148 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION DN | 281 | 179 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA NS UP | 162 | 103 | All SZGR 2.0 genes in this pathway |