Gene Page: MGAT3
Summary ?
| GeneID | 4248 |
| Symbol | MGAT3 |
| Synonyms | GNT-III|GNT3 |
| Description | mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
| Reference | MIM:604621|HGNC:HGNC:7046|Ensembl:ENSG00000128268|HPRD:07058|Vega:OTTHUMG00000030185 |
| Gene type | protein-coding |
| Map location | 22q13.1 |
| Pascal p-value | 5.144E-5 |
| Fetal beta | -0.179 |
| DMG | 1 (# studies) |
| eGene | Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09312336 | 22 | 39884755 | MGAT3 | 1.25E-9 | 0.031 | 1.32E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6001566 | 22 | 39774448 | MGAT3 | ENSG00000128268.11 | 3.19E-6 | 0 | -78901 | gtex_brain_putamen_basal |
| rs6519190 | 22 | 39774525 | MGAT3 | ENSG00000128268.11 | 2.279E-6 | 0 | -78824 | gtex_brain_putamen_basal |
| rs5757647 | 22 | 39775047 | MGAT3 | ENSG00000128268.11 | 5.101E-6 | 0 | -78302 | gtex_brain_putamen_basal |
| rs5757648 | 22 | 39775156 | MGAT3 | ENSG00000128268.11 | 5.088E-6 | 0 | -78193 | gtex_brain_putamen_basal |
| rs9611165 | 22 | 39775250 | MGAT3 | ENSG00000128268.11 | 5.069E-6 | 0 | -78099 | gtex_brain_putamen_basal |
| rs9611166 | 22 | 39775268 | MGAT3 | ENSG00000128268.11 | 5.069E-6 | 0 | -78081 | gtex_brain_putamen_basal |
| rs6001567 | 22 | 39775400 | MGAT3 | ENSG00000128268.11 | 5.069E-6 | 0 | -77949 | gtex_brain_putamen_basal |
| rs6001568 | 22 | 39775786 | MGAT3 | ENSG00000128268.11 | 9.746E-7 | 0 | -77563 | gtex_brain_putamen_basal |
| rs199739333 | 22 | 39777821 | MGAT3 | ENSG00000128268.11 | 4.525E-6 | 0 | -75528 | gtex_brain_putamen_basal |
| rs1010169 | 22 | 39778167 | MGAT3 | ENSG00000128268.11 | 1.888E-6 | 0 | -75182 | gtex_brain_putamen_basal |
| rs1010170 | 22 | 39778327 | MGAT3 | ENSG00000128268.11 | 1.888E-6 | 0 | -75022 | gtex_brain_putamen_basal |
| rs9611169 | 22 | 39783027 | MGAT3 | ENSG00000128268.11 | 3.538E-6 | 0 | -70322 | gtex_brain_putamen_basal |
| rs2413590 | 22 | 39790191 | MGAT3 | ENSG00000128268.11 | 8.085E-7 | 0 | -63158 | gtex_brain_putamen_basal |
| rs5750808 | 22 | 39790987 | MGAT3 | ENSG00000128268.11 | 8.109E-7 | 0 | -62362 | gtex_brain_putamen_basal |
| rs5750809 | 22 | 39791491 | MGAT3 | ENSG00000128268.11 | 5.757E-7 | 0 | -61858 | gtex_brain_putamen_basal |
| rs5750810 | 22 | 39792943 | MGAT3 | ENSG00000128268.11 | 8.159E-7 | 0 | -60406 | gtex_brain_putamen_basal |
| rs5750811 | 22 | 39793066 | MGAT3 | ENSG00000128268.11 | 8.161E-7 | 0 | -60283 | gtex_brain_putamen_basal |
| rs4321460 | 22 | 39793655 | MGAT3 | ENSG00000128268.11 | 4.92E-6 | 0 | -59694 | gtex_brain_putamen_basal |
| rs4386422 | 22 | 39793734 | MGAT3 | ENSG00000128268.11 | 3.853E-6 | 0 | -59615 | gtex_brain_putamen_basal |
| rs5750813 | 22 | 39795228 | MGAT3 | ENSG00000128268.11 | 2.021E-6 | 0 | -58121 | gtex_brain_putamen_basal |
| rs4821892 | 22 | 39795683 | MGAT3 | ENSG00000128268.11 | 1.419E-6 | 0 | -57666 | gtex_brain_putamen_basal |
| rs4821893 | 22 | 39797779 | MGAT3 | ENSG00000128268.11 | 1.438E-6 | 0 | -55570 | gtex_brain_putamen_basal |
| rs5750815 | 22 | 39798449 | MGAT3 | ENSG00000128268.11 | 1.436E-6 | 0 | -54900 | gtex_brain_putamen_basal |
| rs6001582 | 22 | 39806153 | MGAT3 | ENSG00000128268.11 | 2.434E-6 | 0 | -47196 | gtex_brain_putamen_basal |
| rs4821894 | 22 | 39809820 | MGAT3 | ENSG00000128268.11 | 1.444E-6 | 0 | -43529 | gtex_brain_putamen_basal |
| rs5757659 | 22 | 39812409 | MGAT3 | ENSG00000128268.11 | 1.444E-6 | 0 | -40940 | gtex_brain_putamen_basal |
| rs6001588 | 22 | 39819049 | MGAT3 | ENSG00000128268.11 | 1.444E-6 | 0 | -34300 | gtex_brain_putamen_basal |
| rs5757663 | 22 | 39821319 | MGAT3 | ENSG00000128268.11 | 1.726E-6 | 0 | -32030 | gtex_brain_putamen_basal |
| rs5757665 | 22 | 39821641 | MGAT3 | ENSG00000128268.11 | 1.461E-6 | 0 | -31708 | gtex_brain_putamen_basal |
| rs743838 | 22 | 39824707 | MGAT3 | ENSG00000128268.11 | 1.444E-6 | 0 | -28642 | gtex_brain_putamen_basal |
| rs5750828 | 22 | 39835083 | MGAT3 | ENSG00000128268.11 | 1.762E-6 | 0 | -18266 | gtex_brain_putamen_basal |
| rs6001594 | 22 | 39837472 | MGAT3 | ENSG00000128268.11 | 7.107E-7 | 0 | -15877 | gtex_brain_putamen_basal |
| rs2899318 | 22 | 39837625 | MGAT3 | ENSG00000128268.11 | 7.039E-7 | 0 | -15724 | gtex_brain_putamen_basal |
| rs9611176 | 22 | 39838003 | MGAT3 | ENSG00000128268.11 | 1.317E-7 | 0 | -15346 | gtex_brain_putamen_basal |
| rs4821898 | 22 | 39838352 | MGAT3 | ENSG00000128268.11 | 6.727E-7 | 0 | -14997 | gtex_brain_putamen_basal |
| rs5757675 | 22 | 39838892 | MGAT3 | ENSG00000128268.11 | 1.773E-6 | 0 | -14457 | gtex_brain_putamen_basal |
| rs6001595 | 22 | 39839293 | MGAT3 | ENSG00000128268.11 | 4.865E-7 | 0 | -14056 | gtex_brain_putamen_basal |
| rs8138462 | 22 | 39839670 | MGAT3 | ENSG00000128268.11 | 1.735E-6 | 0 | -13679 | gtex_brain_putamen_basal |
| rs5750830 | 22 | 39840828 | MGAT3 | ENSG00000128268.11 | 1.757E-6 | 0 | -12521 | gtex_brain_putamen_basal |
| rs7364148 | 22 | 39842165 | MGAT3 | ENSG00000128268.11 | 1.436E-6 | 0 | -11184 | gtex_brain_putamen_basal |
| rs9306335 | 22 | 39843537 | MGAT3 | ENSG00000128268.11 | 5.318E-6 | 0 | -9812 | gtex_brain_putamen_basal |
| rs8137426 | 22 | 39844350 | MGAT3 | ENSG00000128268.11 | 1.471E-6 | 0 | -8999 | gtex_brain_putamen_basal |
| rs5757680 | 22 | 39844793 | MGAT3 | ENSG00000128268.11 | 2.168E-6 | 0 | -8556 | gtex_brain_putamen_basal |
| rs5757681 | 22 | 39845547 | MGAT3 | ENSG00000128268.11 | 1.095E-6 | 0 | -7802 | gtex_brain_putamen_basal |
| rs1005522 | 22 | 39845898 | MGAT3 | ENSG00000128268.11 | 2.304E-7 | 0 | -7451 | gtex_brain_putamen_basal |
| rs5757682 | 22 | 39848259 | MGAT3 | ENSG00000128268.11 | 1.471E-6 | 0 | -5090 | gtex_brain_putamen_basal |
| rs1557543 | 22 | 39852648 | MGAT3 | ENSG00000128268.11 | 7.285E-7 | 0 | -701 | gtex_brain_putamen_basal |
| rs6001599 | 22 | 39852720 | MGAT3 | ENSG00000128268.11 | 1.471E-6 | 0 | -629 | gtex_brain_putamen_basal |
| rs5995735 | 22 | 39854421 | MGAT3 | ENSG00000128268.11 | 1.095E-6 | 0 | 1072 | gtex_brain_putamen_basal |
| rs5757685 | 22 | 39855540 | MGAT3 | ENSG00000128268.11 | 7.285E-7 | 0 | 2191 | gtex_brain_putamen_basal |
| rs738287 | 22 | 39855728 | MGAT3 | ENSG00000128268.11 | 7.285E-7 | 0 | 2379 | gtex_brain_putamen_basal |
| rs738289 | 22 | 39855883 | MGAT3 | ENSG00000128268.11 | 7.285E-7 | 0 | 2534 | gtex_brain_putamen_basal |
| rs738285 | 22 | 39856356 | MGAT3 | ENSG00000128268.11 | 8.128E-7 | 0 | 3007 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
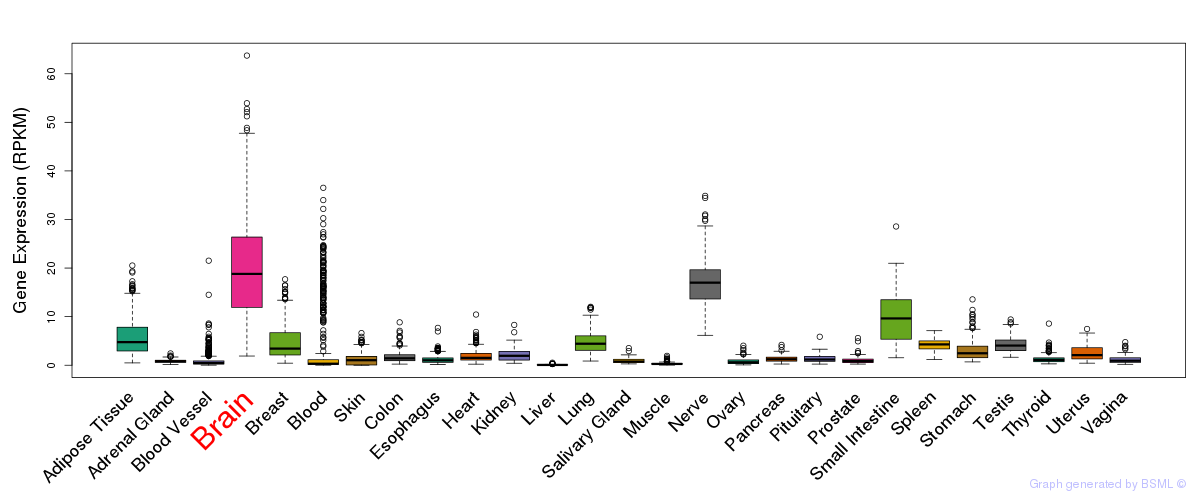
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003830 | beta-1,4-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity | IEA | - | |
| GO:0016757 | transferase activity, transferring glycosyl groups | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006487 | protein amino acid N-linked glycosylation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000139 | Golgi membrane | IEA | - | |
| GO:0005794 | Golgi apparatus | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | TAS | 8370666 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG N GLYCAN BIOSYNTHESIS | 46 | 31 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN STABILIZATION PATHWAY | 42 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| REACTOME POST TRANSLATIONAL PROTEIN MODIFICATION | 188 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME ASPARAGINE N LINKED GLYCOSYLATION | 81 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | 33 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | 18 | 14 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| SUNG METASTASIS STROMA UP | 110 | 70 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 VS CD2 DN | 52 | 35 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| HOFFMAN CLOCK TARGETS DN | 10 | 6 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS DN | 207 | 139 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-504 | 161 | 167 | 1A | hsa-miR-504 | AGACCCUGGUCUGCACUCUAU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.