Gene Page: MYH9
Summary ?
| GeneID | 4627 |
| Symbol | MYH9 |
| Synonyms | BDPLT6|DFNA17|EPSTS|FTNS|MHA|NMHC-II-A|NMMHC-IIA|NMMHCA |
| Description | myosin, heavy chain 9, non-muscle |
| Reference | MIM:160775|HGNC:HGNC:7579|Ensembl:ENSG00000100345|HPRD:01177|Vega:OTTHUMG00000030429 |
| Gene type | protein-coding |
| Map location | 22q13.1 |
| Pascal p-value | 0.27 |
| Sherlock p-value | 0.714 |
| TADA p-value | 0.006 |
| Fetal beta | -0.846 |
| eGene | Myers' cis & trans Meta |
| Support | STRUCTURAL PLASTICITY G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanNRC CompositeSet Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| DNM:Gulsuner_2013 | Whole Exome Sequencing analysis | 155 DNMs identified by exome sequencing of quads or trios of schizophrenia individuals and their parents. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0354 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MYH9 | chr22 | 36689894 | C | T | NM_002473 | p.1285V>M | missense | Schizophrenia | DNM:Fromer_2014 | ||
| MYH9 | chr22 | 36714348 | G | A | NM_002473 | . | silent | Schizophrenia | DNM:Fromer_2014 | ||
| MYH9 | chr22 | 36696896 | C | A | NM_002473 NM_002473 | . . | intronic splice-donor-in2 | Schizophrenia | DNM:Gulsuner_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16827037 | chr3 | 117203284 | MYH9 | 4627 | 0.01 | trans |
Section II. Transcriptome annotation
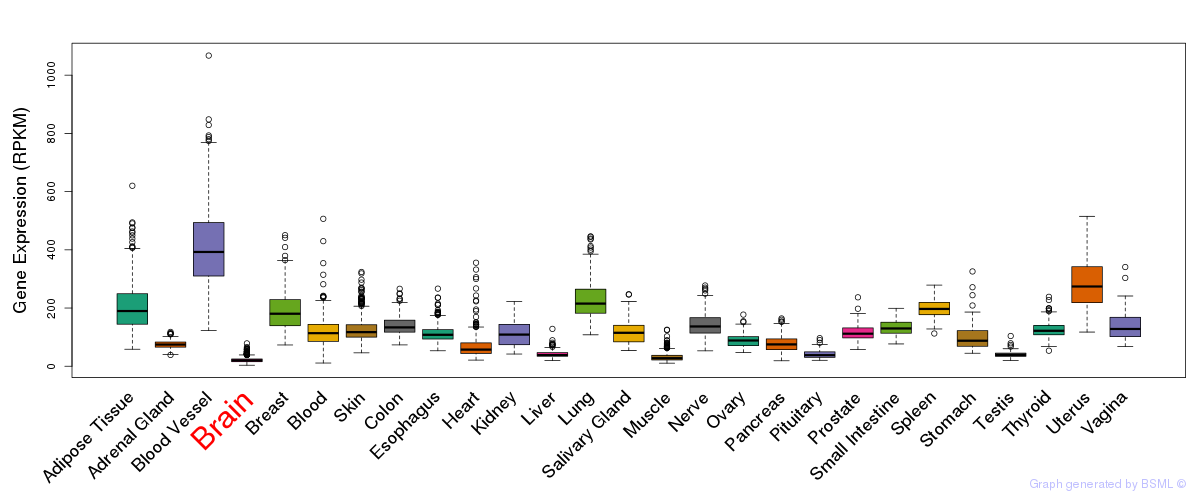
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000146 | microfilament motor activity | IDA | 12237319 | |
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003779 | actin binding | IEA | - | |
| GO:0003774 | motor activity | IEA | - | |
| GO:0005516 | calmodulin binding | IEA | - | |
| GO:0005524 | ATP binding | IDA | 15065866 |15845534 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016887 | ATPase activity | IDA | 12237319 | |
| GO:0043495 | protein anchor | IMP | 16403913 | |
| GO:0051015 | actin filament binding | IDA | 12237319 |14508515 | |
| GO:0051015 | actin filament binding | NAS | 16403913 | |
| GO:0042803 | protein homodimerization activity | IDA | 12237319 | |
| GO:0030898 | actin-dependent ATPase activity | IDA | 15065866 |15845534 | |
| GO:0043531 | ADP binding | IDA | 15065866 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000212 | meiotic spindle organization | IEA | - | |
| GO:0000904 | cell morphogenesis involved in differentiation | IEA | - | |
| GO:0000910 | cytokinesis | IMP | 15774463 | |
| GO:0001525 | angiogenesis | IDA | 16403913 | |
| GO:0001701 | in utero embryonic development | IEA | - | |
| GO:0001768 | establishment of T cell polarity | IEA | - | |
| GO:0030224 | monocyte differentiation | IEP | 1912569 | |
| GO:0007132 | meiotic metaphase I | IEA | - | |
| GO:0006509 | membrane protein ectodomain proteolysis | IDA | 16186248 | |
| GO:0007229 | integrin-mediated signaling pathway | NAS | 10822899 | |
| GO:0008360 | regulation of cell shape | IMP | 11029059 | |
| GO:0007605 | sensory perception of sound | IEA | - | |
| GO:0030048 | actin filament-based movement | IDA | 12237319 | |
| GO:0007520 | myoblast fusion | IEA | - | |
| GO:0006928 | cell motion | IEA | - | |
| GO:0016337 | cell-cell adhesion | IEA | - | |
| GO:0032796 | uropod organization | IEA | - | |
| GO:0015031 | protein transport | IMP | 16403913 | |
| GO:0031532 | actin cytoskeleton reorganization | IMP | 15869600 | |
| GO:0051295 | establishment of meiotic spindle localization | IEA | - | |
| GO:0030220 | platelet formation | IMP | 12237319 | |
| GO:0043534 | blood vessel endothelial cell migration | IMP | 16403913 | |
| GO:0050900 | leukocyte migration | NAS | 12421915 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001772 | immunological synapse | IDA | Synap (GO term level: 7) | 15064761 |
| GO:0031594 | neuromuscular junction | IEA | neuron, axon, Synap, Neurotransmitter (GO term level: 3) | - |
| GO:0001931 | uropod | IDA | Synap (GO term level: 5) | 15064761 |
| GO:0001725 | stress fiber | IDA | 7699007 |14508515 |14706930 |16403913 | |
| GO:0001726 | ruffle | IDA | 16403913 | |
| GO:0005819 | spindle | IEA | - | |
| GO:0005826 | contractile ring | IDA | 11029059 | |
| GO:0005829 | cytosol | IDA | 14508515 | |
| GO:0005634 | nucleus | IDA | 14508515 | |
| GO:0005737 | cytoplasm | IDA | 7699007 | |
| GO:0005938 | cell cortex | IEA | - | |
| GO:0005913 | cell-cell adherens junction | IEA | - | |
| GO:0005886 | plasma membrane | IDA | 16186248 |16403913 | |
| GO:0008305 | integrin complex | IDA | 10822899 | |
| GO:0016459 | myosin complex | IEA | - | |
| GO:0032154 | cleavage furrow | IDA | 7699007 |14508515 | |
| GO:0030863 | cortical cytoskeleton | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG VIRAL MYOCARDITIS | 73 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMA4D IN SEMAPHORIN SIGNALING | 32 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMAPHORIN INTERACTIONS | 68 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | 27 | 21 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP | 285 | 181 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER DN | 169 | 118 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS DN | 73 | 51 | All SZGR 2.0 genes in this pathway |
| BERENJENO ROCK SIGNALING NOT VIA RHOA DN | 48 | 34 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A UP | 191 | 128 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 22 | 13 | 9 | All SZGR 2.0 genes in this pathway |
| XU HGF SIGNALING NOT VIA AKT1 48HR DN | 20 | 12 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 DN | 156 | 106 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A DN | 110 | 78 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A DN | 103 | 71 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN QTL TRANS | 6 | 5 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION DN | 66 | 47 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| XU RESPONSE TO TRETINOIN AND NSC682994 UP | 17 | 13 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 UP | 182 | 119 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 8 | 86 | 57 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 8 HR DN | 53 | 40 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS DN | 96 | 71 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| DAVIES MULTIPLE MYELOMA VS MGUS DN | 28 | 18 | All SZGR 2.0 genes in this pathway |
| ROSS LEUKEMIA WITH MLL FUSIONS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| HOFFMANN SMALL PRE BII TO IMMATURE B LYMPHOCYTE DN | 50 | 33 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 TARGETS UP | 66 | 43 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| MARIADASON RESPONSE TO BUTYRATE CURCUMIN SULINDAC TSA 1 | 10 | 7 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA MESENCHYMAL | 216 | 130 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR133 TARGETS UP | 43 | 27 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 183 | 189 | 1A | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-124.1 | 1297 | 1303 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 1296 | 1303 | 1A,m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-133 | 787 | 794 | 1A,m8 | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| miR-142-3p | 1324 | 1330 | 1A | hsa-miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA |
| miR-149 | 833 | 839 | 1A | hsa-miR-149brain | UCUGGCUCCGUGUCUUCACUCC |
| miR-199 | 1075 | 1081 | m8 | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
| miR-25/32/92/363/367 | 672 | 678 | m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-30-5p | 1339 | 1345 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-410 | 1283 | 1289 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-542-3p | 1317 | 1323 | m8 | hsa-miR-542-3p | UGUGACAGAUUGAUAACUGAAA |
| miR-9 | 226 | 233 | 1A,m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.