Gene Page: NEDD9
Summary ?
| GeneID | 4739 |
| Symbol | NEDD9 |
| Synonyms | CAS-L|CAS2|CASL|CASS2|HEF1 |
| Description | neural precursor cell expressed, developmentally down-regulated 9 |
| Reference | MIM:602265|HGNC:HGNC:7733|Ensembl:ENSG00000111859|HPRD:11888|Vega:OTTHUMG00000014255 |
| Gene type | protein-coding |
| Map location | 6p24.2 |
| Pascal p-value | 0.183 |
| Fetal beta | 2.44 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0159 | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15972264 | 6 | 11279680 | NEDD9 | 4.64E-6 | 0.999 | 0.01 | DMG:Wockner_2014 |
| cg16955407 | 6 | 11279656 | NEDD9 | 6.31E-5 | 0.415 | 0.023 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
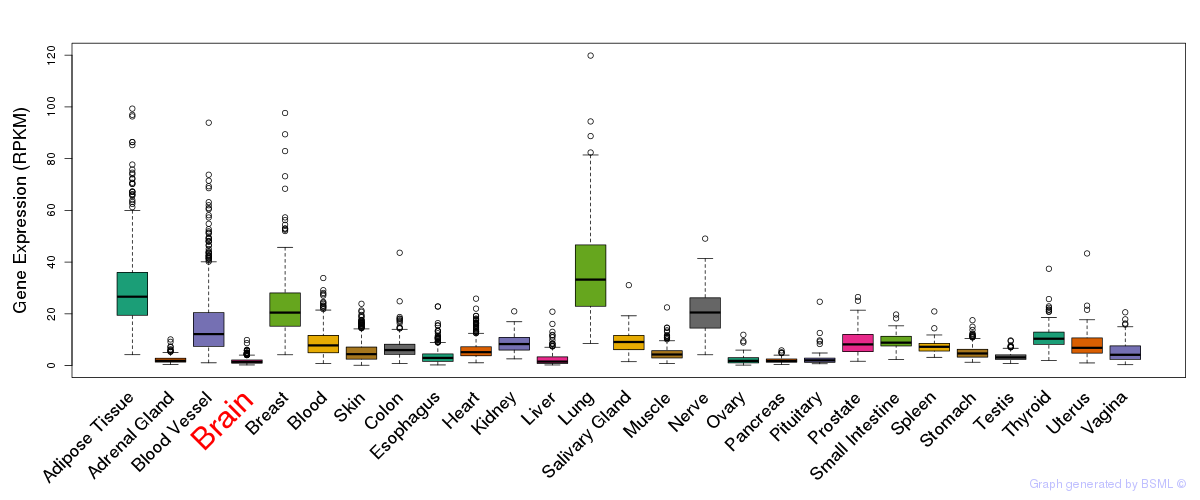
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 11827972 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001558 | regulation of cell growth | IEA | - | |
| GO:0007155 | cell adhesion | NAS | 9584194 | |
| GO:0007229 | integrin-mediated signaling pathway | NAS | 11827972 | |
| GO:0007010 | cytoskeleton organization | NAS | 11827972 | |
| GO:0007165 | signal transduction | TAS | 8668148 | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0007067 | mitosis | IEA | - | |
| GO:0051017 | actin filament bundle formation | NAS | 11827972 | |
| GO:0051301 | cell division | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042995 | cell projection | IEA | axon (GO term level: 4) | - |
| GO:0005794 | Golgi apparatus | IEA | - | |
| GO:0005819 | spindle | TAS | 9584194 | |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005634 | nucleus | TAS | 8668148 | |
| GO:0005737 | cytoplasm | TAS | 9584194 | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | - | HPRD,BioGRID | 8668148 |8879209 |
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | - | HPRD | 8668148 |9366405 |
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | Affinity Capture-Western | BioGRID | 10502414 |
| BCAR3 | KIAA0554 | NSP2 | SH2D3B | breast cancer anti-estrogen resistance 3 | - | HPRD,BioGRID | 12517963 |
| CDH1 | Arc-1 | CD324 | CDHE | ECAD | LCAM | UVO | cadherin 1, type 1, E-cadherin (epithelial) | - | HPRD,BioGRID | 15144564 |
| CHAT | CMS1A | CMS1A2 | choline acetyltransferase | - | HPRD | 10692442 |
| CRK | CRKII | v-crk sarcoma virus CT10 oncogene homolog (avian) | - | HPRD | 8879209 |9366405 |9497377 |
| CRK | CRKII | v-crk sarcoma virus CT10 oncogene homolog (avian) | Affinity Capture-Western Reconstituted Complex | BioGRID | 8879209 |9497377 |
| CRKL | - | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | - | HPRD | 9020138 |9405482 |9498705 |
| CRKL | - | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | Affinity Capture-Western Reconstituted Complex | BioGRID | 9020138 |9162067 |9498705 |9820532 |
| DIMT1L | DIMT1 | HSA9761 | DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) | - | HPRD | 9584194 |
| ELSPBP1 | E12 | HE12 | epididymal sperm binding protein 1 | - | HPRD,BioGRID | 10502414 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD,BioGRID | 9360983 |
| FZR1 | CDC20C | CDH1 | FZR | FZR2 | HCDH | HCDH1 | KIAA1242 | fizzy/cell division cycle 20 related 1 (Drosophila) | CDH1 interacts with HEF1. | BIND | 15144564 |
| ID2 | GIG8 | ID2A | ID2H | MGC26389 | bHLHb26 | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | - | HPRD,BioGRID | 10502414 |
| ITCH | AIF4 | AIP4 | NAPP1 | dJ468O1.1 | itchy E3 ubiquitin protein ligase homolog (mouse) | - | HPRD,BioGRID | 15051726 |
| LCK | YT16 | p56lck | pp58lck | lymphocyte-specific protein tyrosine kinase | - | HPRD,BioGRID | 8879209 |
| LYN | FLJ26625 | JTK8 | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | - | HPRD,BioGRID | 9020138 |
| MICAL1 | DKFZp434B1517 | FLJ11937 | FLJ21739 | MICAL | MICAL-1 | NICAL | microtubule associated monoxygenase, calponin and LIM domain containing 1 | - | HPRD,BioGRID | 11827972 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | - | HPRD,BioGRID | 8879209 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | - | HPRD | 10808124 |
| NEDD9 | CAS-L | CAS2 | CASL | CASS2 | HEF1 | dJ49G10.2 | dJ761I2.1 | neural precursor cell expressed, developmentally down-regulated 9 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 8668148 |10502414 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD,BioGRID | 8668148 |8879209 |10455189 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD,BioGRID | 9020138 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | - | HPRD | 8879209 |
| PTPN12 | PTP-PEST | PTPG1 | protein tyrosine phosphatase, non-receptor type 12 | - | HPRD,BioGRID | 9748319 |
| PXN | FLJ16691 | paxillin | - | HPRD,BioGRID | 10455189 |
| RAPGEF1 | C3G | DKFZp781P1719 | GRF2 | Rap guanine nucleotide exchange factor (GEF) 1 | Affinity Capture-Western | BioGRID | 9498705 |
| SH2D3C | CHAT | FLJ39664 | NSP3 | PRO34088 | SH2 domain containing 3C | Affinity Capture-Western | BioGRID | 10692442 |
| SMAD1 | BSP1 | JV4-1 | JV41 | MADH1 | MADR1 | SMAD family member 1 | - | HPRD,BioGRID | 11118211 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | - | HPRD,BioGRID | 11118211 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | Smad3 interacts with HEF1. | BIND | 15144564 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | - | HPRD,BioGRID | 11118211 |15051726 |15144564 |
| TCF3 | E2A | ITF1 | MGC129647 | MGC129648 | bHLHb21 | transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) | - | HPRD | 10502414 |
| TRIP6 | MGC10556 | MGC10558 | MGC29959 | MGC3837 | MGC4423 | OIP1 | ZRP-1 | thyroid hormone receptor interactor 6 | - | HPRD,BioGRID | 11782456 |
| ZYX | ESP-2 | HED-2 | zyxin | - | HPRD,BioGRID | 11782456 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP | 185 | 126 | All SZGR 2.0 genes in this pathway |
| SCIBETTA KDM5B TARGETS DN | 81 | 55 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP | 176 | 123 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA EGF SIGNALING UP | 58 | 40 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN | 209 | 122 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 DN | 153 | 100 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION UP | 211 | 136 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS C UP | 170 | 114 | All SZGR 2.0 genes in this pathway |
| WANG BARRETTS ESOPHAGUS AND ESOPHAGUS CANCER DN | 37 | 22 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| MAHADEVAN GIST MORPHOLOGICAL SWITCH | 15 | 11 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM1 | 229 | 137 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 DN | 156 | 106 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A UP | 142 | 104 | All SZGR 2.0 genes in this pathway |
| SCIAN CELL CYCLE TARGETS OF TP53 AND TP73 UP | 9 | 7 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| WU CELL MIGRATION | 184 | 114 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE DN | 165 | 104 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 HELA | 164 | 118 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 120 MCF10A | 65 | 44 | All SZGR 2.0 genes in this pathway |
| JI METASTASIS REPRESSED BY STK11 | 27 | 17 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC TGFA UP | 61 | 40 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA UP | 207 | 145 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 9 | 92 | 59 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN | 220 | 147 | All SZGR 2.0 genes in this pathway |
| JI CARCINOGENESIS BY KRAS AND STK11 DN | 17 | 12 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN | 216 | 143 | All SZGR 2.0 genes in this pathway |
| CHEN HOXA5 TARGETS 9HR UP | 223 | 132 | All SZGR 2.0 genes in this pathway |
| JIANG TIP30 TARGETS UP | 46 | 28 | All SZGR 2.0 genes in this pathway |
| BOHN PRIMARY IMMUNODEFICIENCY SYNDROM DN | 40 | 31 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A4 | 196 | 124 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| UZONYI RESPONSE TO LEUKOTRIENE AND THROMBIN | 37 | 26 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 UP | 134 | 85 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| ISSAEVA MLL2 TARGETS | 62 | 35 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA NS UP | 162 | 103 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 360 | 367 | 1A,m8 | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-125/351 | 153 | 159 | 1A | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-145 | 47 | 54 | 1A,m8 | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-18 | 338 | 345 | 1A,m8 | hsa-miR-18a | UAAGGUGCAUCUAGUGCAGAUA |
| hsa-miR-18b | UAAGGUGCAUCUAGUGCAGUUA | ||||
| miR-24 | 436 | 442 | m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.