Gene Page: OPCML
Summary ?
| GeneID | 4978 |
| Symbol | OPCML |
| Synonyms | IGLON1|OBCAM|OPCM |
| Description | opioid binding protein/cell adhesion molecule-like |
| Reference | MIM:600632|HGNC:HGNC:8143|Ensembl:ENSG00000183715|HPRD:07198|Vega:OTTHUMG00000163658 |
| Gene type | protein-coding |
| Map location | 11q25 |
| Pascal p-value | 2.958E-4 |
| Sherlock p-value | 0.124 |
| DMG | 2 (# studies) |
| eGene | Myers' cis & trans |
| Support | CELL ADHESION AND TRANSSYNAPTIC SIGNALING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_clathrin G2Cdb.human_Synaptosome CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 31 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02224864 | 11 | 132934554 | OPCML | 1.081E-4 | -0.408 | 0.028 | DMG:Wockner_2014 |
| cg02338757 | 11 | 132814117 | OPCML | 1.51E-9 | -0.011 | 1.41E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4366501 | chr11 | 133380324 | OPCML | 4978 | 0.11 | cis | ||
| rs7708940 | chr5 | 150789050 | OPCML | 4978 | 0.12 | trans |
Section II. Transcriptome annotation
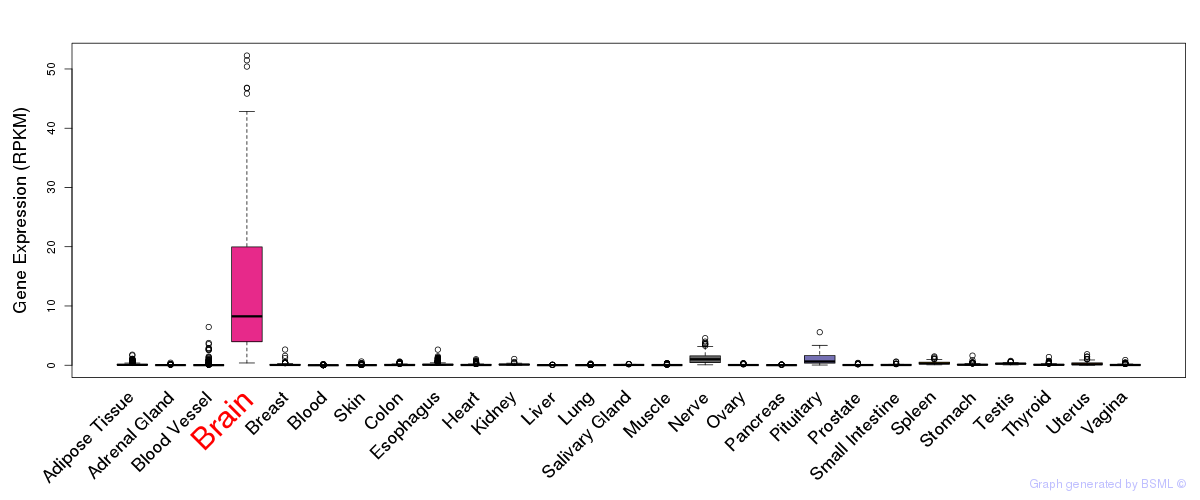
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MAG | 0.95 | 0.90 |
| CNP | 0.95 | 0.87 |
| FA2H | 0.95 | 0.90 |
| KLK6 | 0.93 | 0.88 |
| ADAMTS4 | 0.93 | 0.91 |
| PLP1 | 0.93 | 0.89 |
| GJC2 | 0.93 | 0.86 |
| PMP22 | 0.93 | 0.90 |
| HSPA2 | 0.92 | 0.88 |
| TF | 0.92 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TRNAU1AP | -0.53 | -0.66 |
| NKIRAS2 | -0.53 | -0.60 |
| TUBB2B | -0.52 | -0.71 |
| NR2C2AP | -0.52 | -0.68 |
| KIAA1949 | -0.51 | -0.63 |
| AIM2 | -0.51 | -0.73 |
| YBX1 | -0.51 | -0.68 |
| HN1 | -0.51 | -0.59 |
| ZNF821 | -0.50 | -0.61 |
| IFT52 | -0.50 | -0.65 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| GO:0004985 | opioid receptor activity | TAS | 1333602 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008038 | neuron recognition | TAS | neuron (GO term level: 4) | 7891157 |
| GO:0007155 | cell adhesion | TAS | 7721093 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005886 | plasma membrane | TAS | 1333602 | |
| GO:0005887 | integral to plasma membrane | TAS | 1333602 | |
| GO:0031225 | anchored to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| LOPES METHYLATED IN COLON CANCER UP | 27 | 20 | All SZGR 2.0 genes in this pathway |
| HARRIS BRAIN CANCER PROGENITORS | 44 | 23 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF ICP WITH H3K4ME3 AND H3K27ME3 | 38 | 34 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS ICP WITH H3K4ME3 AND H327ME3 | 126 | 83 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 AND H3K27ME3 | 137 | 85 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| GOBERT CORE OLIGODENDROCYTE DIFFERENTIATION | 40 | 28 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL UP | 105 | 46 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-134 | 1209 | 1215 | m8 | hsa-miR-134brain | UGUGACUGGUUGACCAGAGGG |
| miR-190 | 39 | 45 | 1A | hsa-miR-190 | UGAUAUGUUUGAUAUAUUAGGU |
| miR-194 | 5120 | 5126 | 1A | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| miR-196 | 5101 | 5107 | m8 | hsa-miR-196a | UAGGUAGUUUCAUGUUGUUGG |
| hsa-miR-196b | UAGGUAGUUUCCUGUUGUUGG | ||||
| miR-224 | 1630 | 1636 | 1A | hsa-miR-224 | CAAGUCACUAGUGGUUCCGUUUA |
| miR-376c | 4942 | 4948 | m8 | hsa-miR-376c | AACAUAGAGGAAAUUCCACG |
| miR-379 | 203 | 209 | 1A | hsa-miR-379brain | UGGUAGACUAUGGAACGUA |
| miR-452 | 1665 | 1672 | 1A,m8 | hsa-miR-452 | UGUUUGCAGAGGAAACUGAGAC |
| hsa-miR-452 | UGUUUGCAGAGGAAACUGAGAC | ||||
| miR-494 | 1589 | 1595 | 1A | hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU |
| miR-9 | 3242 | 3248 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 1644 | 1650 | m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.