Gene Page: OPHN1
Summary ?
| GeneID | 4983 |
| Symbol | OPHN1 |
| Synonyms | ARHGAP41|MRX60|OPN1 |
| Description | oligophrenin 1 |
| Reference | MIM:300127|HGNC:HGNC:8148|Ensembl:ENSG00000079482|HPRD:02130|Vega:OTTHUMG00000021744 |
| Gene type | protein-coding |
| Map location | Xq12 |
| Fetal beta | 0.55 |
| eGene | Myers' cis & trans |
| Support | Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2768571 | chrX | 67258888 | OPHN1 | 4983 | 0.14 | cis | ||
| rs4852316 | chr2 | 73919657 | OPHN1 | 4983 | 0.13 | trans | ||
| rs17350056 | chr2 | 73922876 | OPHN1 | 4983 | 0.18 | trans | ||
| rs12052539 | chr2 | 73937152 | OPHN1 | 4983 | 0.12 | trans | ||
| rs17350188 | chr2 | 73964841 | OPHN1 | 4983 | 0.1 | trans | ||
| rs12624267 | chr2 | 73986930 | OPHN1 | 4983 | 0.14 | trans | ||
| rs9875049 | chr3 | 56457469 | OPHN1 | 4983 | 0.09 | trans | ||
| rs1665659 | chr10 | 118448297 | OPHN1 | 4983 | 0.18 | trans |
Section II. Transcriptome annotation
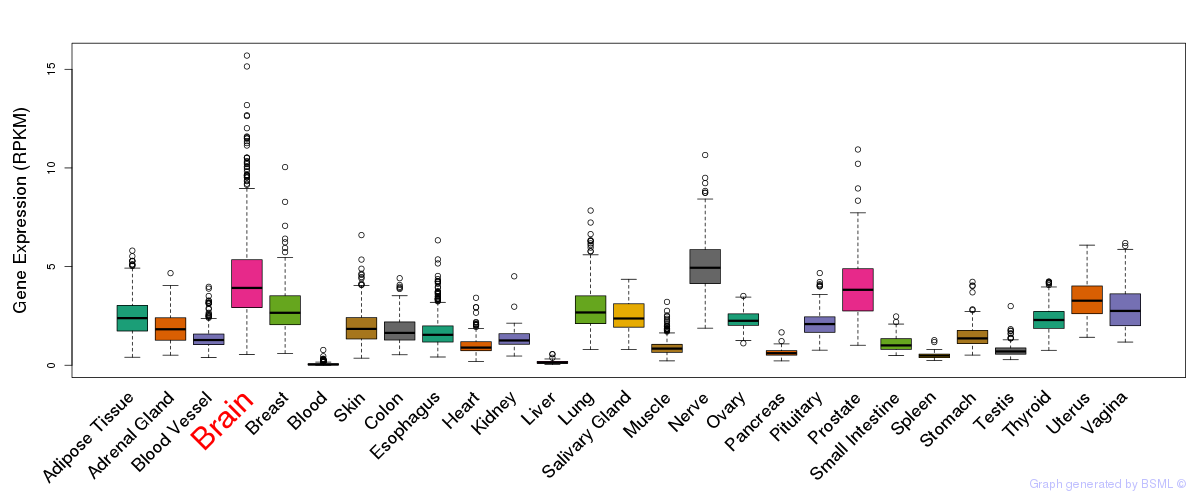
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TAL1 | 0.87 | 0.16 |
| MAB21L1 | 0.85 | 0.48 |
| KLK10 | 0.84 | 0.21 |
| ANO2 | 0.84 | 0.33 |
| SP5 | 0.83 | 0.27 |
| MC4R | 0.81 | 0.17 |
| MAP4K1 | 0.80 | 0.26 |
| LEF1 | 0.79 | 0.21 |
| LMO1 | 0.78 | 0.41 |
| CEACAM21 | 0.78 | 0.18 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLC26A4 | -0.22 | -0.30 |
| BAIAP2 | -0.22 | -0.22 |
| CACNG3 | -0.20 | -0.41 |
| C20orf39 | -0.20 | -0.15 |
| C1orf115 | -0.19 | -0.29 |
| KIAA1614 | -0.19 | 0.01 |
| GTDC1 | -0.19 | -0.01 |
| PIK3IP1 | -0.19 | -0.24 |
| SPARCL1 | -0.19 | -0.52 |
| KCNJ4 | -0.18 | -0.39 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005100 | Rho GTPase activator activity | TAS | 9582072 | |
| GO:0003779 | actin binding | IEA | - | |
| GO:0005096 | GTPase activator activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007411 | axon guidance | TAS | axon (GO term level: 13) | 9582072 |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 9582072 |
| GO:0007165 | signal transduction | TAS | 9582072 | |
| GO:0006930 | substrate-bound cell migration, cell extension | TAS | 9582072 | |
| GO:0030036 | actin cytoskeleton organization | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0005886 | plasma membrane | IDA | 18029348 | |
| GO:0015629 | actin cytoskeleton | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA RHO PATHWAY | 32 | 23 | All SZGR 2.0 genes in this pathway |
| PID RHOA REG PATHWAY | 46 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY RHO GTPASES | 113 | 81 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| MATTIOLI MGUS VS PCL | 116 | 62 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| CHANG POU5F1 TARGETS UP | 15 | 10 | All SZGR 2.0 genes in this pathway |
| STOSSI RESPONSE TO ESTRADIOL | 50 | 35 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP | 268 | 157 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 UP | 295 | 149 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN DN | 150 | 99 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |