Gene Page: SHANK1
Summary ?
| GeneID | 50944 |
| Symbol | SHANK1 |
| Synonyms | SPANK-1|SSTRIP|synamon |
| Description | SH3 and multiple ankyrin repeat domains 1 |
| Reference | MIM:604999|HGNC:HGNC:15474|Ensembl:ENSG00000161681|HPRD:05413|Vega:OTTHUMG00000137380 |
| Gene type | protein-coding |
| Map location | 19q13.3 |
| Pascal p-value | 0.055 |
| TADA p-value | 0.005 |
| Fetal beta | -0.196 |
| DMG | 1 (# studies) |
| eGene | Meta |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_mGluR5 G2Cdb.humanNRC CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0923 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SHANK1 | chr19 | 51171670 | T | TG | NM_016148 | . | frameshift | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17826344 | 19 | 51169660 | SHANK1 | 7.29E-5 | -0.271 | 0.025 | DMG:Wockner_2014 |
| cg04005969 | 19 | 51171247 | SHANK1 | 2.258E-4 | -0.345 | 0.036 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
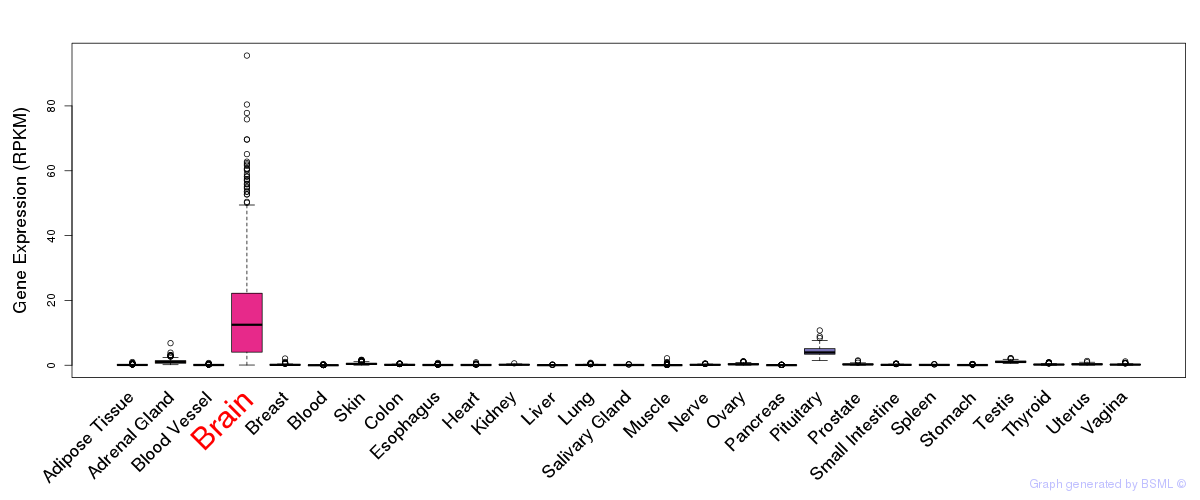
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 10551867 |11583995 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007016 | cytoskeletal anchoring at plasma membrane | NAS | 10551867 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0030425 | dendrite | NAS | neuron, axon, dendrite (GO term level: 6) | 10551867 |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005622 | intracellular | NAS | 10551867 | |
| GO:0005624 | membrane fraction | IDA | 10551867 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARHGEF7 | BETA-PIX | COOL1 | DKFZp686C12170 | DKFZp761K1021 | KIAA0142 | KIAA0412 | Nbla10314 | P50 | P50BP | P85 | P85COOL1 | P85SPR | PAK3 | PIXB | Rho guanine nucleotide exchange factor (GEF) 7 | - | HPRD,BioGRID | 12626503 |
| BAI2 | - | brain-specific angiogenesis inhibitor 2 | - | HPRD,BioGRID | 10964907 |
| BAIAP2 | BAP2 | IRSP53 | BAI1-associated protein 2 | - | HPRD,BioGRID | 12504591 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | - | HPRD,BioGRID | 10433268 |
| DLGAP1 | DAP-1 | DAP-1-ALPHA | DAP-1-BETA | GKAP | MGC88156 | SAPAP1 | hGKAP | discs, large (Drosophila) homolog-associated protein 1 | Co-crystal Structure | BioGRID | 12954649 |
| DNM2 | CMTDI1 | CMTDIB | DI-CMTB | DYN2 | DYNII | dynamin 2 | - | HPRD,BioGRID | 11583995 |
| LPHN1 | CIRL1 | CL1 | LEC2 | latrophilin 1 | CIRL1 interacts with SSTRIP. This interaction was modeled on a demonstrated interaction between CIRL1 from an unspecified species and human SSTRIP . | BIND | 10964907 |
| LPHN1 | CIRL1 | CL1 | LEC2 | latrophilin 1 | Two-hybrid | BioGRID | 10964907 |
| LPHN2 | CIRL2 | CL2 | LEC1 | LPHH1 | latrophilin 2 | Two-hybrid | BioGRID | 10964907 |
| SHANK1 | SPANK-1 | SSTRIP | synamon | SH3 and multiple ankyrin repeat domains 1 | - | HPRD,BioGRID | 12954649 |
| SHARPIN | DKFZp434N1923 | SIPL1 | SHANK-associated RH domain interactor | - | HPRD,BioGRID | 11178875 |12753155 |
| SPTAN1 | (ALPHA)II-SPECTRIN | FLJ44613 | NEAS | spectrin, alpha, non-erythrocytic 1 (alpha-fodrin) | - | HPRD,BioGRID | 11509555 |
| SSTR2 | - | somatostatin receptor 2 | - | HPRD,BioGRID | 10551867 |
| TANC1 | KIAA1728 | ROLSB | TANC | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 | - | HPRD | 15673434 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-184 | 24 | 30 | 1A | hsa-miR-184 | UGGACGGAGAACUGAUAAGGGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.