Gene Page: SERPINA5
Summary ?
| GeneID | 5104 |
| Symbol | SERPINA5 |
| Synonyms | PAI-3|PAI3|PCI|PCI-B|PLANH3|PROCI |
| Description | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| Reference | MIM:601841|HGNC:HGNC:8723|Ensembl:ENSG00000188488|HPRD:03503|Vega:OTTHUMG00000170860 |
| Gene type | protein-coding |
| Map location | 14q32.1 |
| Pascal p-value | 0.194 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:McCarthy_2014 | Whole Exome Sequencing analysis | Whole exome sequencing of 57 trios with sporadic or familial schizophrenia. | |
| Expression | Meta-analysis of gene expression | P value: 2.04 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SERPINA5 | chr14 | 95058502 | G | A | NM_000624 | p.V383M | missense SNV | N | B | Schizophrenia | DNM:McCarthy_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7268640 | chr20 | 47150267 | SERPINA5 | 5104 | 2.902E-4 | trans |
Section II. Transcriptome annotation
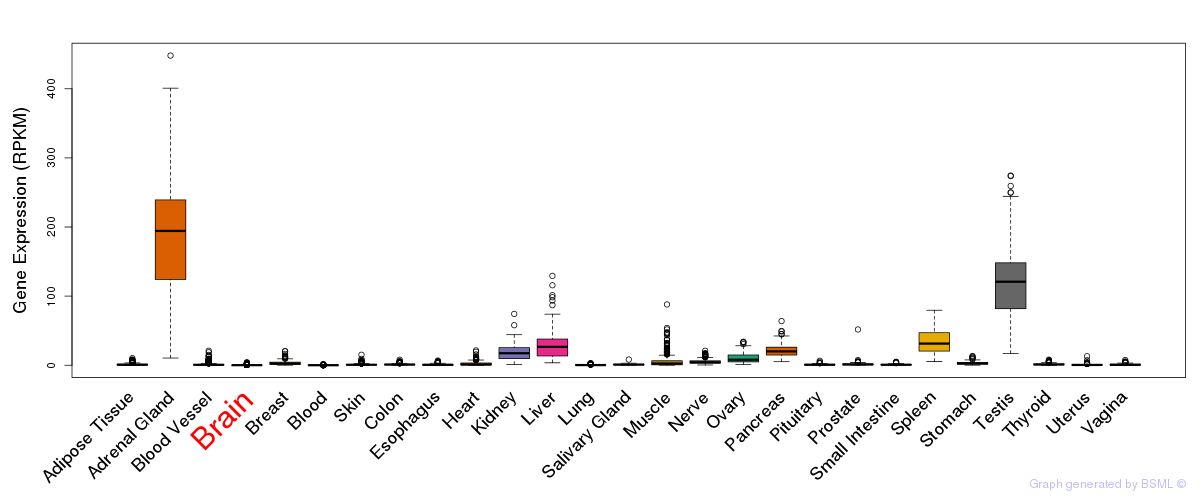
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| STT3B | 0.87 | 0.72 |
| DOK6 | 0.86 | 0.82 |
| ARSK | 0.84 | 0.71 |
| ST6GALNAC3 | 0.83 | 0.72 |
| LCORL | 0.82 | 0.65 |
| VRK1 | 0.82 | 0.65 |
| HCRTR2 | 0.81 | 0.58 |
| C14orf23 | 0.81 | 0.58 |
| RNF138 | 0.80 | 0.65 |
| C5orf23 | 0.80 | 0.68 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HSD17B14 | -0.46 | -0.59 |
| MT-CO2 | -0.46 | -0.60 |
| SLC7A10 | -0.46 | -0.66 |
| S100A16 | -0.46 | -0.60 |
| RAB34 | -0.46 | -0.59 |
| VEGFB | -0.46 | -0.50 |
| C19orf36 | -0.46 | -0.54 |
| LIMS2 | -0.46 | -0.50 |
| MT-CYB | -0.45 | -0.59 |
| PARP10 | -0.45 | -0.45 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004867 | serine-type endopeptidase inhibitor activity | IDA | 7521127 | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0008201 | heparin binding | TAS | 11120760 | |
| GO:0032190 | acrosin binding | IPI | 7521127 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007283 | spermatogenesis | ISS | - | |
| GO:0007342 | fusion of sperm to egg plasma membrane | NAS | 9510955 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | NAS | 14718574 | |
| GO:0005576 | extracellular region | TAS | 11120760 | |
| GO:0016020 | membrane | IDA | 7521127 | |
| GO:0043234 | protein complex | IDA | 7521127 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACR | - | acrosin | - | HPRD,BioGRID | 7521127 |
| COPS6 | CSN6 | MOV34-34KD | COP9 constitutive photomorphogenic homolog subunit 6 (Arabidopsis) | Two-hybrid | BioGRID | 16169070 |
| F10 | FX | FXA | coagulation factor X | - | HPRD | 2551064 |
| F11 | FXI | MGC141891 | coagulation factor XI | - | HPRD,BioGRID | 2551064 |
| F2 | PT | coagulation factor II (thrombin) | - | HPRD | 12878585 |
| FGA | Fib2 | MGC119422 | MGC119423 | MGC119425 | fibrinogen alpha chain | - | HPRD,BioGRID | 8384496 |8589203 |
| FGB | MGC104327 | MGC120405 | fibrinogen beta chain | - | HPRD,BioGRID | 8148485 |8384496 |
| FGG | - | fibrinogen gamma chain | - | HPRD | 8384496 |
| KLK1 | KLKR | Klk6 | hK1 | kallikrein 1 | - | HPRD | 9368023 |
| KLK2 | KLK2A2 | MGC12201 | hK2 | kallikrein-related peptidase 2 | Affinity Capture-Western | BioGRID | 9016396 |
| KLK2 | KLK2A2 | MGC12201 | hK2 | kallikrein-related peptidase 2 | - | HPRD | 9368023 |
| KLK3 | APS | KLK2A1 | PSA | hK3 | kallikrein-related peptidase 3 | - | HPRD,BioGRID | 7509746 |8665956 |8732755 |
| KLKB1 | KLK3 | PPK | kallikrein B, plasma (Fletcher factor) 1 | - | HPRD,BioGRID | 2551064 |11864713 |
| PLAT | DKFZp686I03148 | T-PA | TPA | plasminogen activator, tissue | - | HPRD,BioGRID | 2551064 |
| PLAU | ATF | UPA | URK | UROKINASE | u-PA | plasminogen activator, urokinase | Reconstituted Complex | BioGRID | 2551064 |2752144 |
| PLAU | ATF | UPA | URK | UROKINASE | u-PA | plasminogen activator, urokinase | - | HPRD | 2752144 |9365930 |
| PROC | PC | PROC1 | protein C (inactivator of coagulation factors Va and VIIIa) | - | HPRD,BioGRID | 11123896 |
| SEMG2 | SGII | semenogelin II | - | HPRD | 8665956 |
| THBD | CD141 | THRM | TM | thrombomodulin | - | HPRD | 12878585 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG COMPLEMENT AND COAGULATION CASCADES | 69 | 49 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 UP | 112 | 72 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| NIELSEN SCHWANNOMA UP | 18 | 16 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS DN | 142 | 94 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN | 220 | 147 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| MASSARWEH RESPONSE TO ESTRADIOL | 61 | 47 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE DN | 274 | 165 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BRAIN DN | 85 | 58 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE UP | 97 | 61 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP | 172 | 109 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| MATZUK MALE REPRODUCTION SERTOLI | 28 | 23 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH DN | 31 | 25 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER DN | 134 | 83 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| NABA ECM REGULATORS | 238 | 125 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |