| KEGG PPAR SIGNALING PATHWAY
| 69 | 47 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY
| 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME PPARA ACTIVATES GENE EXPRESSION
| 104 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS
| 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM
| 168 | 115 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION
| 72 | 53 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA UP
| 92 | 57 | All SZGR 2.0 genes in this pathway |
| HOOI ST7 TARGETS DN
| 123 | 78 | All SZGR 2.0 genes in this pathway |
| CORRE MULTIPLE MYELOMA UP
| 74 | 45 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP
| 544 | 308 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA UP
| 171 | 112 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA BY DMOG UP
| 130 | 85 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A TARGETS DN
| 91 | 58 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A AND HIF2A TARGETS DN
| 104 | 72 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP
| 293 | 179 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL DN
| 186 | 107 | All SZGR 2.0 genes in this pathway |
| XU HGF SIGNALING NOT VIA AKT1 6HR
| 26 | 15 | All SZGR 2.0 genes in this pathway |
| XU HGF TARGETS REPRESSED BY AKT1 UP
| 10 | 7 | All SZGR 2.0 genes in this pathway |
| YORDY RECIPROCAL REGULATION BY ETS1 AND SP100 DN
| 87 | 48 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL UP
| 83 | 51 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A UP
| 142 | 104 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP
| 1037 | 673 | All SZGR 2.0 genes in this pathway |
| INGRAM SHH TARGETS DN
| 64 | 41 | All SZGR 2.0 genes in this pathway |
| WOTTON RUNX TARGETS UP
| 21 | 14 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 240 MCF10A
| 57 | 36 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA UP
| 207 | 145 | All SZGR 2.0 genes in this pathway |
| GOLDRATH IMMUNE MEMORY
| 65 | 42 | All SZGR 2.0 genes in this pathway |
| GERY CEBP TARGETS
| 126 | 90 | All SZGR 2.0 genes in this pathway |
| MENSE HYPOXIA UP
| 98 | 71 | All SZGR 2.0 genes in this pathway |
| KIM HYPOXIA
| 25 | 21 | All SZGR 2.0 genes in this pathway |
| MCDOWELL ACUTE LUNG INJURY UP
| 45 | 29 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 4
| 48 | 32 | All SZGR 2.0 genes in this pathway |
| LIU SMARCA4 TARGETS
| 64 | 39 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL
| 311 | 205 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE
| 261 | 166 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED
| 1022 | 619 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN
| 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP
| 648 | 398 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP
| 425 | 298 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE
| 242 | 168 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN
| 138 | 92 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C UP
| 47 | 29 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA DN
| 267 | 160 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA2 DN
| 80 | 51 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 11
| 103 | 68 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP
| 857 | 456 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE
| 1137 | 655 | All SZGR 2.0 genes in this pathway |
| KIM GLIS2 TARGETS UP
| 84 | 61 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP
| 682 | 433 | All SZGR 2.0 genes in this pathway |
| FARDIN HYPOXIA 11
| 32 | 29 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7
| 403 | 240 | All SZGR 2.0 genes in this pathway |
| PEDERSEN TARGETS OF 611CTF ISOFORM OF ERBB2
| 76 | 45 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D
| 882 | 506 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP
| 364 | 236 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A
| 770 | 480 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS UP
| 221 | 120 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN
| 308 | 187 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP
| 297 | 194 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF
| 516 | 308 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS
| 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED
| 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME
| 1028 | 559 | All SZGR 2.0 genes in this pathway |
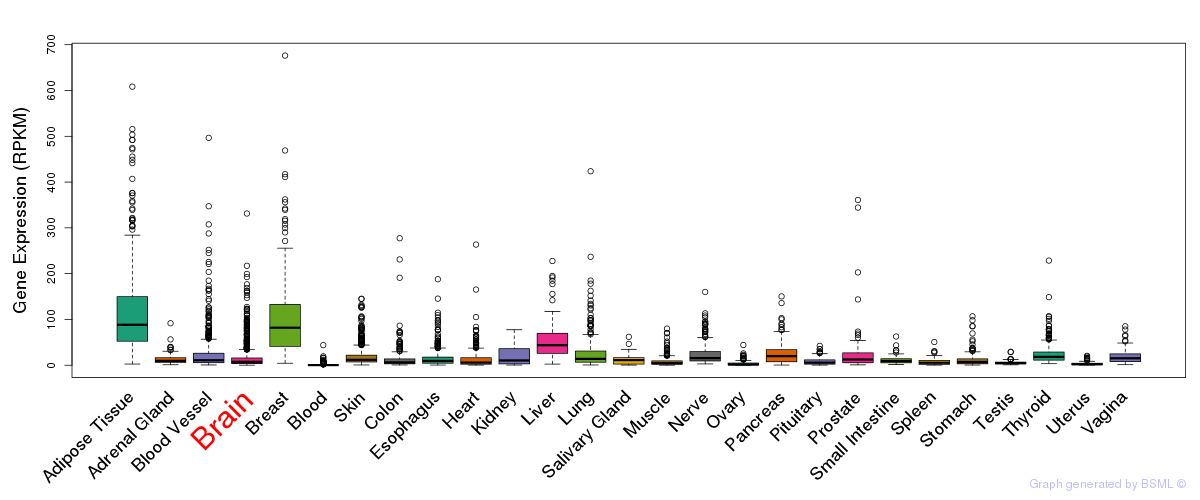
 eQTL annotation
eQTL annotation