Gene Page: JKAMP
Summary ?
| GeneID | 51528 |
| Symbol | JKAMP |
| Synonyms | C14orf100|CDA06|HSPC213|HSPC327|JAMP |
| Description | JNK1/MAPK8-associated membrane protein |
| Reference | MIM:611176|HGNC:HGNC:20184|Ensembl:ENSG00000050130|HPRD:12623| |
| Gene type | protein-coding |
| Map location | 14q23.1 |
| Pascal p-value | 0.005 |
| Sherlock p-value | 8.849E-5 |
| Fetal beta | 0.39 |
| eGene | Hippocampus Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1253165 | chr14 | 59883137 | JKAMP | 51528 | 0.01 | cis | ||
| rs1271183 | chr14 | 59891790 | JKAMP | 51528 | 0.01 | cis | ||
| rs17834074 | chr14 | 59891982 | JKAMP | 51528 | 0.04 | cis | ||
| rs1253170 | chr14 | 59893646 | JKAMP | 51528 | 0.01 | cis | ||
| rs1273156 | chr14 | 59899933 | JKAMP | 51528 | 0.01 | cis | ||
| rs2774052 | chr14 | 59900019 | JKAMP | 51528 | 0.01 | cis | ||
| rs1262135 | chr14 | 59909767 | JKAMP | 51528 | 0.01 | cis | ||
| rs10136708 | chr14 | 59927375 | JKAMP | 51528 | 2.522E-6 | cis | ||
| rs8660 | chr14 | 59939726 | JKAMP | 51528 | 4.243E-4 | cis | ||
| rs1253103 | chr14 | 59949447 | JKAMP | 51528 | 7.45E-7 | cis | ||
| rs7151295 | chr14 | 59978071 | JKAMP | 51528 | 2.338E-5 | cis | ||
| rs6573270 | chr14 | 60029166 | JKAMP | 51528 | 3.51E-4 | cis | ||
| rs4901941 | chr14 | 60031644 | JKAMP | 51528 | 1.683E-6 | cis | ||
| rs17255933 | chr14 | 60042938 | JKAMP | 51528 | 0 | cis | ||
| rs7143940 | chr14 | 60047544 | JKAMP | 51528 | 0 | cis | ||
| rs11158264 | chr14 | 60050255 | JKAMP | 51528 | 0.03 | cis | ||
| rs7157903 | chr14 | 60051459 | JKAMP | 51528 | 0 | cis | ||
| rs10139045 | chr14 | 60060572 | JKAMP | 51528 | 0.01 | cis | ||
| rs473689 | chr11 | 103740691 | JKAMP | 51528 | 0.07 | trans | ||
| rs10136708 | chr14 | 59927375 | JKAMP | 51528 | 4.347E-4 | trans | ||
| rs8660 | chr14 | 59939726 | JKAMP | 51528 | 0.03 | trans | ||
| rs1253103 | chr14 | 59949447 | JKAMP | 51528 | 1.423E-4 | trans | ||
| rs7151295 | chr14 | 59978071 | JKAMP | 51528 | 0 | trans | ||
| rs6573270 | chr14 | 60029166 | JKAMP | 51528 | 0.03 | trans | ||
| rs4901941 | chr14 | 60031644 | JKAMP | 51528 | 3.065E-4 | trans | ||
| rs17255933 | chr14 | 60042938 | JKAMP | 51528 | 0.17 | trans | ||
| rs7143940 | chr14 | 60047544 | JKAMP | 51528 | 0.14 | trans | ||
| rs7157903 | chr14 | 60051459 | JKAMP | 51528 | 0.18 | trans |
Section II. Transcriptome annotation
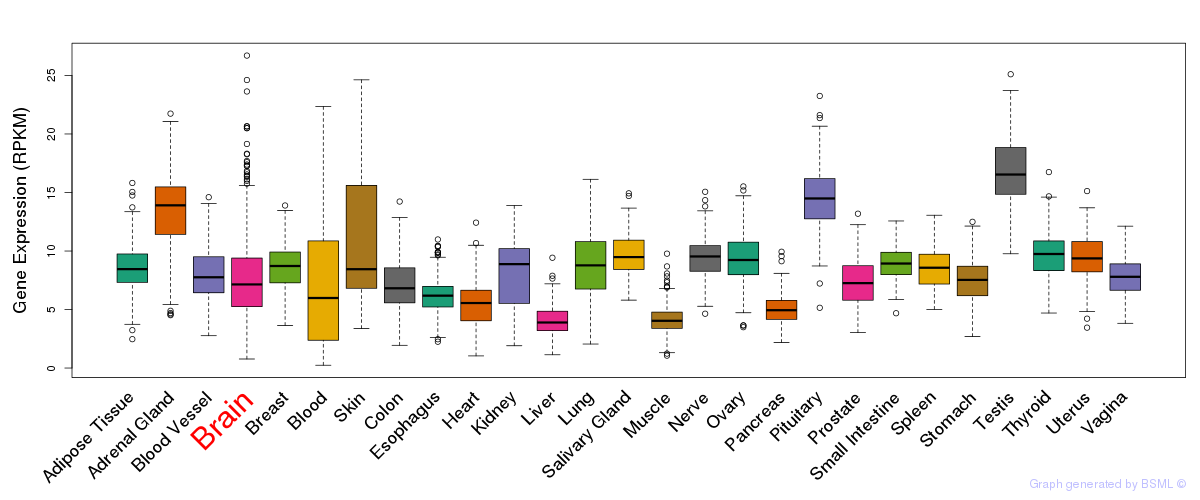
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| LIANG HEMATOPOIESIS STEM CELL NUMBER LARGE VS TINY UP | 44 | 24 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| GUO HEX TARGETS DN | 65 | 36 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS DN | 314 | 188 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| MOLENAAR TARGETS OF CCND1 AND CDK4 DN | 58 | 25 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A | 182 | 108 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-182 | 17 | 23 | 1A | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-410 | 541 | 547 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-96 | 17 | 23 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.