Gene Page: PDZK1
Summary ?
| GeneID | 5174 |
| Symbol | PDZK1 |
| Synonyms | CAP70|CLAMP|NHERF-3|NHERF3|PDZD1 |
| Description | PDZ domain containing 1 |
| Reference | MIM:603831|HGNC:HGNC:8821|Ensembl:ENSG00000174827|HPRD:04829|Vega:OTTHUMG00000013735 |
| Gene type | protein-coding |
| Map location | 1q21 |
| Fetal beta | -0.435 |
| eGene | Cerebellum Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1934584 | chr1 | 245971711 | PDZK1 | 5174 | 0.05 | trans | ||
| rs16829545 | chr2 | 151977407 | PDZK1 | 5174 | 1.663E-5 | trans | ||
| rs7584986 | chr2 | 184111432 | PDZK1 | 5174 | 0.04 | trans | ||
| rs16878852 | chr4 | 27115684 | PDZK1 | 5174 | 0.01 | trans | ||
| rs17762315 | chr5 | 76807576 | PDZK1 | 5174 | 0.1 | trans | ||
| rs504430 | chr12 | 129477364 | PDZK1 | 5174 | 0.18 | trans | ||
| rs10847737 | chr12 | 129507602 | PDZK1 | 5174 | 0.07 | trans | ||
| rs16955618 | chr15 | 29937543 | PDZK1 | 5174 | 9.479E-5 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
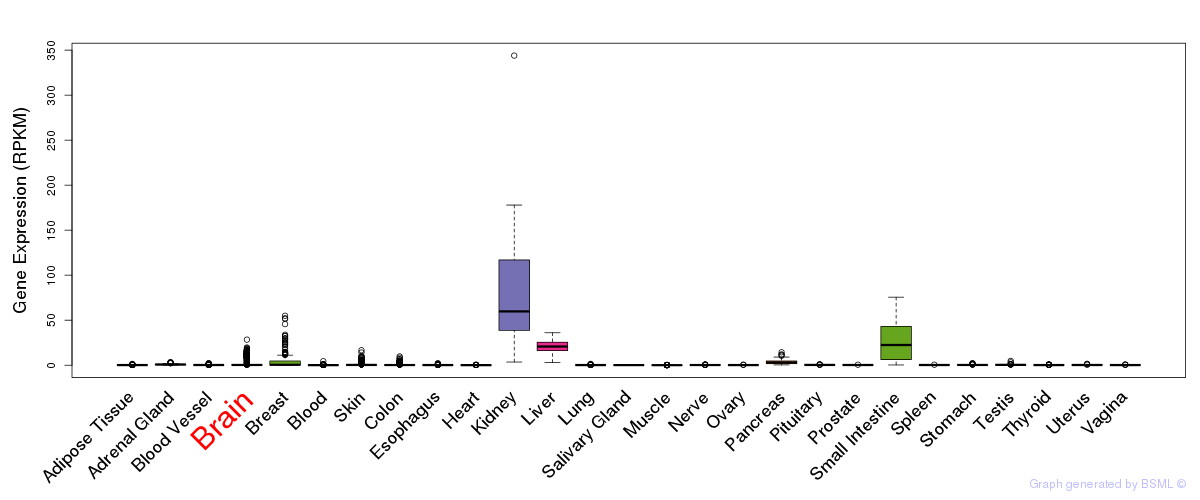
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005215 | transporter activity | TAS | 9461128 | |
| GO:0030165 | PDZ domain binding | IPI | 16236806 |16738539 | |
| GO:0008022 | protein C-terminus binding | IPI | 15304510 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008283 | cell proliferation | TAS | 9461128 | |
| GO:0015893 | drug transport | NAS | 16738539 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0016020 | membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABCC2 | ABC30 | CMOAT | DJS | KIAA1010 | MRP2 | cMRP | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 | - | HPRD,BioGRID | 10496535 |12615054 |
| AKAP10 | D-AKAP2 | MGC9414 | PRKA10 | A kinase (PRKA) anchor protein 10 | - | HPRD,BioGRID | 14531806 |
| CFTR | ABC35 | ABCC7 | CF | CFTR/MRP | MRP7 | TNR-CFTR | dJ760C5.1 | cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) | - | HPRD,BioGRID | 11051556 |12471024 |12615054 |
| CLCN3 | CLC3 | ClC-3 | chloride channel 3 | - | HPRD,BioGRID | 12471024 |
| FARP2 | FIR | FRG | KIAA0793 | PLEKHC3 | FERM, RhoGEF and pleckstrin domain protein 2 | Reconstituted Complex Two-hybrid | BioGRID | 14531806 |
| GFAP | FLJ45472 | glial fibrillary acidic protein | Two-hybrid | BioGRID | 16189514 |
| PDZK1IP1 | DD96 | MAP17 | RP1-18D14.5 | SPAP | PDZK1 interacting protein 1 | - | HPRD,BioGRID | 9461128 |12754212 |12837682 |14531806 |
| PEX5 | FLJ50634 | FLJ50721 | FLJ51948 | PTS1-BP | PTS1R | PXR1 | peroxisomal biogenesis factor 5 | Two-hybrid | BioGRID | 16189514 |
| SCARB1 | CD36L1 | CLA-1 | CLA1 | MGC138242 | SR-BI | SRB1 | scavenger receptor class B, member 1 | - | HPRD,BioGRID | 10829064 |12119305 |14551195 |
| SLC17A1 | MGC126794 | MGC126796 | NAPI-1 | NPT-1 | NPT1 | solute carrier family 17 (sodium phosphate), member 1 | - | HPRD,BioGRID | 14531806 |
| SLC22A12 | OAT4L | RST | URAT1 | solute carrier family 22 (organic anion/urate transporter), member 12 | - | HPRD,BioGRID | 14531806 |
| SLC22A4 | MGC34546 | MGC40524 | OCTN1 | solute carrier family 22 (organic cation/ergothioneine transporter), member 4 | Reconstituted Complex Two-hybrid | BioGRID | 14531806 |
| SLC34A3 | FLJ38680 | HHRH | NPTIIc | solute carrier family 34 (sodium phosphate), member 3 | Reconstituted Complex Two-hybrid | BioGRID | 14531806 |
| SLC9A3 | MGC126718 | MGC126720 | NHE3 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 | - | HPRD,BioGRID | 14531806 |
| SLC9A3R1 | EBP50 | NHERF | NHERF1 | NPHLOP2 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 | Reconstituted Complex Two-hybrid | BioGRID | 14531806 |
| SLC9A3R2 | E3KARP | MGC104639 | NHE3RF2 | NHERF-2 | NHERF2 | OCTS2 | SIP-1 | SIP1 | TKA-1 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 | Two-hybrid | BioGRID | 14531806 |
| SLK | KIAA0204 | LOSK | MGC133067 | STK2 | bA16H23.1 | se20-9 | STE20-like kinase (yeast) | Reconstituted Complex Two-hybrid | BioGRID | 14531806 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | Two-hybrid | BioGRID | 14531806 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA MTA3 PATHWAY | 19 | 10 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS DUCTAL NORMAL DN | 198 | 110 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS LOBULAR NORMAL DN | 69 | 43 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS DUCTAL NORMAL DN | 91 | 53 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS LOBULAR NORMAL UP | 94 | 59 | All SZGR 2.0 genes in this pathway |
| SAMOLS TARGETS OF KHSV MIRNAS DN | 62 | 35 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 UP | 112 | 72 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP | 372 | 227 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 6HR DN | 167 | 100 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA DN | 146 | 94 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA BY DMOG DN | 59 | 40 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| KORKOLA YOLK SAC TUMOR | 62 | 33 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO ESTRADIOL UP | 37 | 28 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| BECKER TAMOXIFEN RESISTANCE DN | 52 | 37 | All SZGR 2.0 genes in this pathway |
| STOSSI RESPONSE TO ESTRADIOL | 50 | 35 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA CANCER | 83 | 52 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| MASSARWEH RESPONSE TO ESTRADIOL | 61 | 47 | All SZGR 2.0 genes in this pathway |
| MASRI RESISTANCE TO TAMOXIFEN AND AROMATASE INHIBITORS UP | 20 | 10 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING | 246 | 152 | All SZGR 2.0 genes in this pathway |
| CADWELL ATG16L1 TARGETS UP | 93 | 56 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER DN | 134 | 83 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| HOELZEL NF1 TARGETS UP | 139 | 93 | All SZGR 2.0 genes in this pathway |
| OHGUCHI LIVER HNF4A TARGETS DN | 149 | 85 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 UP | 211 | 131 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP C | 92 | 60 | All SZGR 2.0 genes in this pathway |
| VANDESLUIS COMMD1 TARGETS GROUP 3 UP | 89 | 50 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |