Gene Page: PITX3
Summary ?
| GeneID | 5309 |
| Symbol | PITX3 |
| Synonyms | ASMD|ASOD|CTPP4|CTRCT11|PTX3 |
| Description | paired like homeodomain 3 |
| Reference | MIM:602669|HGNC:HGNC:9006|Ensembl:ENSG00000107859|HPRD:04051|Vega:OTTHUMG00000018952 |
| Gene type | protein-coding |
| Map location | 10q24.32 |
| Pascal p-value | 0.028 |
| Fetal beta | -0.255 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
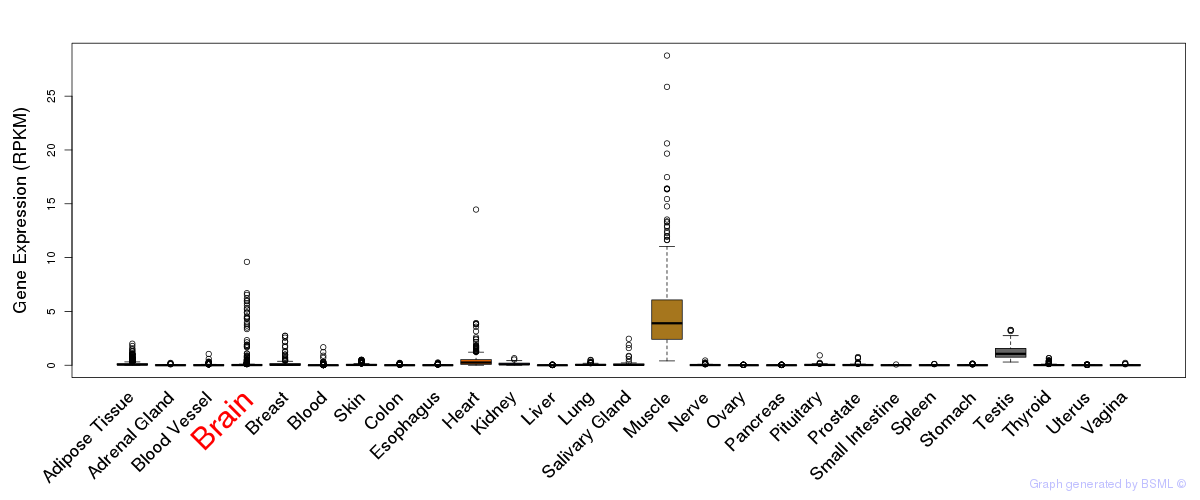
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CYBB | 0.78 | 0.76 |
| LCP1 | 0.75 | 0.70 |
| DOCK2 | 0.75 | 0.71 |
| ITGB2 | 0.74 | 0.70 |
| NCKAP1L | 0.74 | 0.68 |
| TLR7 | 0.74 | 0.66 |
| SYK | 0.73 | 0.67 |
| CSF1R | 0.72 | 0.70 |
| IRF8 | 0.72 | 0.67 |
| LAPTM5 | 0.72 | 0.68 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.18 | -0.29 | -0.28 |
| AF347015.2 | -0.28 | -0.27 |
| MT-CO2 | -0.27 | -0.28 |
| AF347015.21 | -0.27 | -0.22 |
| AF347015.31 | -0.26 | -0.27 |
| AF347015.26 | -0.25 | -0.24 |
| AF347015.8 | -0.25 | -0.25 |
| AC100783.1 | -0.25 | -0.27 |
| IL32 | -0.24 | -0.20 |
| CXCL14 | -0.24 | -0.19 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | IEA | - | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0048666 | neuron development | IEA | neuron (GO term level: 9) | - |
| GO:0030901 | midbrain development | IEA | Brain (GO term level: 8) | - |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0009887 | organ morphogenesis | TAS | 9620774 | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP | 268 | 157 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| DAVIES MULTIPLE MYELOMA VS MGUS UP | 13 | 10 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR DN | 88 | 59 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D | 280 | 158 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-133 | 220 | 226 | m8 | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| miR-328 | 236 | 242 | 1A | hsa-miR-328brain | CUGGCCCUCUCUGCCCUUCCGU |
| miR-339 | 217 | 223 | 1A | hsa-miR-339 | UCCCUGUCCUCCAGGAGCUCA |
| miR-542-5p | 20 | 27 | 1A,m8 | hsa-miR-542-5p | UCGGGGAUCAUCAUGUCACGAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.