Gene Page: MBD3
Summary ?
| GeneID | 53615 |
| Symbol | MBD3 |
| Synonyms | - |
| Description | methyl-CpG binding domain protein 3 |
| Reference | MIM:603573|HGNC:HGNC:6918|Ensembl:ENSG00000071655|HPRD:04653|Vega:OTTHUMG00000180081 |
| Gene type | protein-coding |
| Map location | 19p13.3 |
| Pascal p-value | 0.089 |
| Sherlock p-value | 0.363 |
| Fetal beta | -0.133 |
| eGene | Myers' cis & trans |
| Support | Ascano FMRP targets Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4491879 | chr3 | 189373908 | MBD3 | 53615 | 0.16 | trans | ||
| rs17542473 | chr4 | 111182451 | MBD3 | 53615 | 0.2 | trans | ||
| rs2143256 | chr20 | 9664538 | MBD3 | 53615 | 0.09 | trans | ||
| rs6039548 | chr20 | 9666092 | MBD3 | 53615 | 0.14 | trans | ||
| rs6056802 | 0 | MBD3 | 53615 | 0.09 | trans |
Section II. Transcriptome annotation
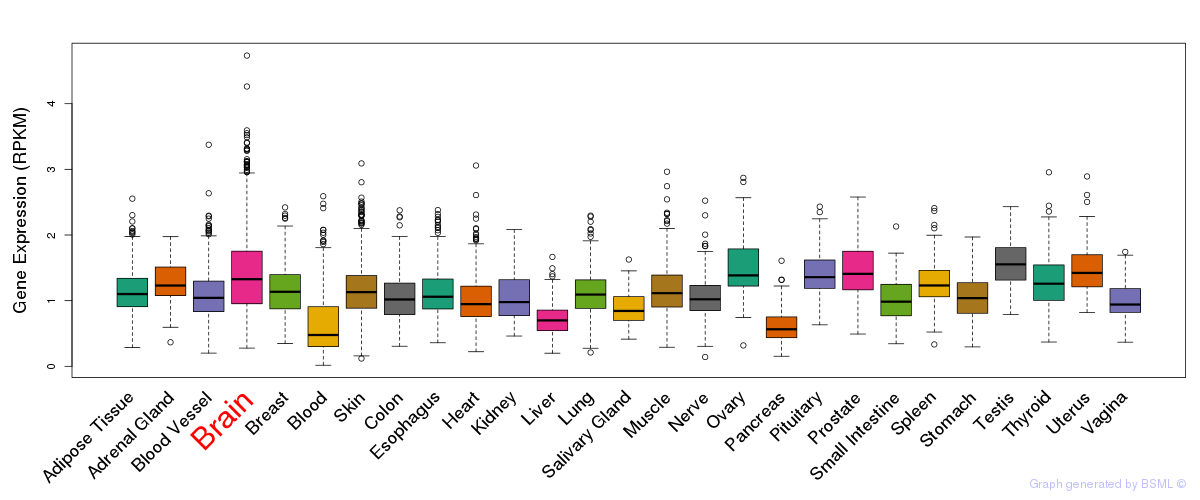
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AURKA | AIK | ARK1 | AURA | AURORA2 | BTAK | MGC34538 | STK15 | STK6 | STK7 | aurora kinase A | - | HPRD,BioGRID | 12354758 |
| BCL6 | BCL5 | BCL6A | LAZ3 | ZBTB27 | ZNF51 | B-cell CLL/lymphoma 6 | BCL6 interacts with MBD3. | BIND | 15454082 |
| CHD4 | DKFZp686E06161 | Mi-2b | Mi2-BETA | chromodomain helicase DNA binding protein 4 | Affinity Capture-Western | BioGRID | 12354758 |
| DNMT1 | AIM | CXXC9 | DNMT | FLJ16293 | MCMT | MGC104992 | DNA (cytosine-5-)-methyltransferase 1 | - | HPRD | 10947852 |
| GATAD2A | FLJ20085 | FLJ21017 | p66alpha | GATA zinc finger domain containing 2A | - | HPRD,BioGRID | 12183469 |
| GATAD2B | FLJ37346 | KIAA1150 | MGC138257 | MGC138285 | P66beta | RP11-216N14.6 | GATA zinc finger domain containing 2B | - | HPRD | 12183469 |
| GATAD2B | FLJ37346 | KIAA1150 | MGC138257 | MGC138285 | P66beta | RP11-216N14.6 | GATA zinc finger domain containing 2B | - | HPRD,BioGRID | 11756549 |12183469 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD,BioGRID | 12124384 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | MBD3 interacts with HDAC1. | BIND | 12124384 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | Affinity Capture-Western | BioGRID | 12354758 |
| MBD2 | DKFZp586O0821 | DMTase | NY-CO-41 | methyl-CpG binding domain protein 2 | - | HPRD | 10947852 |
| MBD2 | DKFZp586O0821 | DMTase | NY-CO-41 | methyl-CpG binding domain protein 2 | Reconstituted Complex Two-hybrid | BioGRID | 10444591 |15456747 |
| MBD3 | - | methyl-CpG binding domain protein 3 | Two-hybrid | BioGRID | 15456747 |
| MBD3L1 | MBD3L | MGC138263 | MGC138269 | methyl-CpG binding domain protein 3-like 1 | - | HPRD,BioGRID | 15456747 |
| MTA2 | DKFZp686F2281 | MTA1L1 | PID | metastasis associated 1 family, member 2 | MBD3 interacts with MTA2. | BIND | 12124384 |
| MTA2 | DKFZp686F2281 | MTA1L1 | PID | metastasis associated 1 family, member 2 | - | HPRD,BioGRID | 12124384 |
| RBBP4 | NURF55 | RBAP48 | retinoblastoma binding protein 4 | - | HPRD,BioGRID | 10444591 |
| RBBP7 | MGC138867 | MGC138868 | RbAp46 | retinoblastoma binding protein 7 | - | HPRD,BioGRID | 10444591 |
| SPEN | KIAA0929 | MINT | RP1-134O19.1 | SHARP | spen homolog, transcriptional regulator (Drosophila) | - | HPRD | 11331609 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA MTA3 PATHWAY | 19 | 10 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSI PATHWAY | 66 | 50 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| MOOTHA VOXPHOS | 87 | 51 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE UP | 157 | 105 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC | 210 | 123 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 2D DN | 64 | 35 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE DN | 274 | 165 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |