Gene Page: KIF1A
Summary ?
| GeneID | 547 |
| Symbol | KIF1A |
| Synonyms | ATSV|C2orf20|HSN2C|MRD9|SPG30|UNC104 |
| Description | kinesin family member 1A |
| Reference | MIM:601255|HGNC:HGNC:888|Ensembl:ENSG00000130294|HPRD:03156|Vega:OTTHUMG00000151940 |
| Gene type | protein-coding |
| Map location | 2q37.3 |
| Pascal p-value | 0.618 |
| DMG | 1 (# studies) |
| Support | INTRACELLULAR TRAFFICKING CompositeSet Darnell FMRP targets Ascano FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| KIF1A | chr2 | 241658510 | G | T | NM_001244008 NM_004321 | p.1709N>K p.1608N>K | missense missense | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03896836 | 2 | 241760025 | KIF1A | 8.89E-6 | -0.493 | 0.013 | DMG:Wockner_2014 |
| cg17500202 | 2 | 241722099 | KIF1A | 1.107E-4 | 0.514 | 0.028 | DMG:Wockner_2014 |
| cg27378591 | 2 | 241697024 | KIF1A | 4.164E-4 | 0.513 | 0.044 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
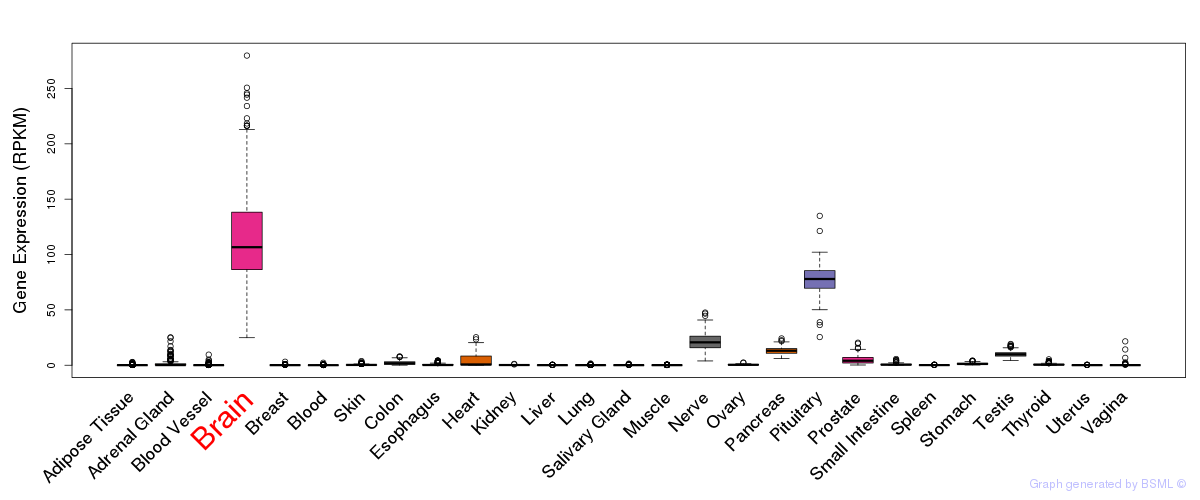
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TUSC4 | 0.91 | 0.89 |
| CHKB | 0.87 | 0.87 |
| COQ6 | 0.87 | 0.84 |
| GEMIN7 | 0.84 | 0.80 |
| COMMD4 | 0.84 | 0.84 |
| MPDU1 | 0.83 | 0.80 |
| RNF25 | 0.83 | 0.84 |
| HDHD3 | 0.83 | 0.78 |
| OVCA2 | 0.83 | 0.86 |
| MTX1 | 0.83 | 0.78 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-ATP8 | -0.52 | -0.63 |
| AF347015.8 | -0.51 | -0.56 |
| AF347015.18 | -0.50 | -0.60 |
| AF347015.26 | -0.50 | -0.58 |
| MT-CYB | -0.49 | -0.55 |
| AF347015.2 | -0.49 | -0.55 |
| AF347015.15 | -0.49 | -0.54 |
| AF347015.27 | -0.49 | -0.56 |
| PTRF | -0.47 | -0.49 |
| MT-CO2 | -0.46 | -0.51 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003777 | microtubule motor activity | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008089 | anterograde axon cargo transport | TAS | axon (GO term level: 9) | 7539720 |
| GO:0007018 | microtubule-based movement | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005794 | Golgi apparatus | IDA | 18029348 | |
| GO:0005874 | microtubule | IEA | - | |
| GO:0005875 | microtubule associated complex | IEA | - | |
| GO:0005634 | nucleus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IDA | 18029348 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| WONG ENDMETRIUM CANCER UP | 25 | 17 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE UP | 134 | 93 | All SZGR 2.0 genes in this pathway |
| LEE TARGETS OF PTCH1 AND SUFU DN | 83 | 69 | All SZGR 2.0 genes in this pathway |
| LIAN LIPA TARGETS 6M | 74 | 47 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 DN | 65 | 45 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP | 221 | 150 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS UP | 108 | 78 | All SZGR 2.0 genes in this pathway |
| NIELSEN SYNOVIAL SARCOMA UP | 18 | 14 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 3535 | 3541 | m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-141/200a | 3515 | 3521 | 1A | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.