Gene Page: LEPROT
Summary ?
| GeneID | 54741 |
| Symbol | LEPROT |
| Synonyms | LEPR|OB-RGRP|OBRGRP|VPS55 |
| Description | leptin receptor overlapping transcript |
| Reference | MIM:613461|HGNC:HGNC:29477|Ensembl:ENSG00000213625|HPRD:13987|Vega:OTTHUMG00000009065Vega:OTTHUMG00000009115 |
| Gene type | protein-coding |
| Map location | 1p31.3 |
| Sherlock p-value | 0.844 |
| Fetal beta | 0.093 |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cortex Frontal Cortex BA9 Nucleus accumbens basal ganglia Putamen basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.02692 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs71058403 | 1 | 65834062 | LEPROT | ENSG00000213625.4 | 1.338E-6 | 0.01 | -59403 | gtex_brain_ba24 |
| rs9326063 | 1 | 65835911 | LEPROT | ENSG00000213625.4 | 1.833E-6 | 0.01 | -57554 | gtex_brain_ba24 |
| rs10158872 | 1 | 65836230 | LEPROT | ENSG00000213625.4 | 1.839E-6 | 0.01 | -57235 | gtex_brain_ba24 |
| rs10158873 | 1 | 65836244 | LEPROT | ENSG00000213625.4 | 1.839E-6 | 0.01 | -57221 | gtex_brain_ba24 |
| rs11208637 | 1 | 65836553 | LEPROT | ENSG00000213625.4 | 1.817E-6 | 0.01 | -56912 | gtex_brain_ba24 |
| rs8179492 | 1 | 65836908 | LEPROT | ENSG00000213625.4 | 1.839E-6 | 0.01 | -56557 | gtex_brain_ba24 |
| rs6588136 | 1 | 65837119 | LEPROT | ENSG00000213625.4 | 1.839E-6 | 0.01 | -56346 | gtex_brain_ba24 |
| rs6588137 | 1 | 65837282 | LEPROT | ENSG00000213625.4 | 1.839E-6 | 0.01 | -56183 | gtex_brain_ba24 |
| rs6693498 | 1 | 65837389 | LEPROT | ENSG00000213625.4 | 1.839E-6 | 0.01 | -56076 | gtex_brain_ba24 |
| rs7537733 | 1 | 65838229 | LEPROT | ENSG00000213625.4 | 1.839E-6 | 0.01 | -55236 | gtex_brain_ba24 |
| rs6588138 | 1 | 65838366 | LEPROT | ENSG00000213625.4 | 1.92E-6 | 0.01 | -55099 | gtex_brain_ba24 |
| rs6659883 | 1 | 65838618 | LEPROT | ENSG00000213625.4 | 1.793E-6 | 0.01 | -54847 | gtex_brain_ba24 |
| rs1591364 | 1 | 65842364 | LEPROT | ENSG00000213625.4 | 1.701E-6 | 0.01 | -51101 | gtex_brain_ba24 |
| rs716592 | 1 | 65843982 | LEPROT | ENSG00000213625.4 | 1.697E-6 | 0.01 | -49483 | gtex_brain_ba24 |
| rs3818449 | 1 | 65844963 | LEPROT | ENSG00000213625.4 | 1.159E-6 | 0.01 | -48502 | gtex_brain_ba24 |
| rs1334879 | 1 | 65846051 | LEPROT | ENSG00000213625.4 | 7.667E-7 | 0.01 | -47414 | gtex_brain_ba24 |
| rs1334878 | 1 | 65846124 | LEPROT | ENSG00000213625.4 | 5.864E-7 | 0.01 | -47341 | gtex_brain_ba24 |
| rs1334877 | 1 | 65846406 | LEPROT | ENSG00000213625.4 | 7.453E-7 | 0.01 | -47059 | gtex_brain_ba24 |
| rs201227181 | 1 | 65846602 | LEPROT | ENSG00000213625.4 | 6.202E-7 | 0.01 | -46863 | gtex_brain_ba24 |
| rs11208640 | 1 | 65846996 | LEPROT | ENSG00000213625.4 | 7.61E-7 | 0.01 | -46469 | gtex_brain_ba24 |
| rs9728544 | 1 | 65847105 | LEPROT | ENSG00000213625.4 | 7.411E-7 | 0.01 | -46360 | gtex_brain_ba24 |
| rs6697416 | 1 | 65848075 | LEPROT | ENSG00000213625.4 | 7.504E-7 | 0.01 | -45390 | gtex_brain_ba24 |
| rs11208642 | 1 | 65848622 | LEPROT | ENSG00000213625.4 | 7.462E-7 | 0.01 | -44843 | gtex_brain_ba24 |
| rs4593866 | 1 | 65850968 | LEPROT | ENSG00000213625.4 | 7.265E-7 | 0.01 | -42497 | gtex_brain_ba24 |
| rs10889546 | 1 | 65851759 | LEPROT | ENSG00000213625.4 | 7.203E-7 | 0.01 | -41706 | gtex_brain_ba24 |
| rs6700226 | 1 | 65852064 | LEPROT | ENSG00000213625.4 | 7.203E-7 | 0.01 | -41401 | gtex_brain_ba24 |
| rs9436725 | 1 | 65852870 | LEPROT | ENSG00000213625.4 | 7.203E-7 | 0.01 | -40595 | gtex_brain_ba24 |
| rs9436726 | 1 | 65853196 | LEPROT | ENSG00000213625.4 | 7.203E-7 | 0.01 | -40269 | gtex_brain_ba24 |
| rs6588142 | 1 | 65854473 | LEPROT | ENSG00000213625.4 | 7.203E-7 | 0.01 | -38992 | gtex_brain_ba24 |
| rs6588143 | 1 | 65854556 | LEPROT | ENSG00000213625.4 | 7.203E-7 | 0.01 | -38909 | gtex_brain_ba24 |
| rs28429763 | 1 | 65855954 | LEPROT | ENSG00000213625.4 | 1.258E-6 | 0.01 | -37511 | gtex_brain_ba24 |
| rs7545397 | 1 | 65856282 | LEPROT | ENSG00000213625.4 | 7.876E-7 | 0.01 | -37183 | gtex_brain_ba24 |
| rs7545720 | 1 | 65856443 | LEPROT | ENSG00000213625.4 | 7.195E-7 | 0.01 | -37022 | gtex_brain_ba24 |
| rs10789183 | 1 | 65856779 | LEPROT | ENSG00000213625.4 | 7.203E-7 | 0.01 | -36686 | gtex_brain_ba24 |
| rs4554758 | 1 | 65857403 | LEPROT | ENSG00000213625.4 | 1.738E-7 | 0.01 | -36062 | gtex_brain_ba24 |
| rs4575118 | 1 | 65857457 | LEPROT | ENSG00000213625.4 | 1.738E-7 | 0.01 | -36008 | gtex_brain_ba24 |
| rs4512686 | 1 | 65857483 | LEPROT | ENSG00000213625.4 | 1.738E-7 | 0.01 | -35982 | gtex_brain_ba24 |
| rs6588144 | 1 | 65858088 | LEPROT | ENSG00000213625.4 | 1.738E-7 | 0.01 | -35377 | gtex_brain_ba24 |
| rs4325172 | 1 | 65858145 | LEPROT | ENSG00000213625.4 | 3.265E-7 | 0.01 | -35320 | gtex_brain_ba24 |
| rs4332387 | 1 | 65859108 | LEPROT | ENSG00000213625.4 | 1.738E-7 | 0.01 | -34357 | gtex_brain_ba24 |
| rs6588145 | 1 | 65859784 | LEPROT | ENSG00000213625.4 | 6.17E-7 | 0.01 | -33681 | gtex_brain_ba24 |
| rs4582839 | 1 | 65860687 | LEPROT | ENSG00000213625.4 | 4.148E-7 | 0.01 | -32778 | gtex_brain_ba24 |
| rs4329545 | 1 | 65861060 | LEPROT | ENSG00000213625.4 | 7.778E-7 | 0.01 | -32405 | gtex_brain_ba24 |
| rs11208645 | 1 | 65861798 | LEPROT | ENSG00000213625.4 | 3.265E-7 | 0.01 | -31667 | gtex_brain_ba24 |
| rs9436287 | 1 | 65863302 | LEPROT | ENSG00000213625.4 | 3.265E-7 | 0.01 | -30163 | gtex_brain_ba24 |
| rs36067635 | 1 | 65865574 | LEPROT | ENSG00000213625.4 | 1.873E-7 | 0.01 | -27891 | gtex_brain_ba24 |
| rs35782988 | 1 | 65865590 | LEPROT | ENSG00000213625.4 | 3.435E-7 | 0.01 | -27875 | gtex_brain_ba24 |
| rs12726933 | 1 | 65865635 | LEPROT | ENSG00000213625.4 | 3.265E-7 | 0.01 | -27830 | gtex_brain_ba24 |
| rs4916052 | 1 | 65865940 | LEPROT | ENSG00000213625.4 | 1.746E-6 | 0.01 | -27525 | gtex_brain_ba24 |
| rs7524932 | 1 | 65865993 | LEPROT | ENSG00000213625.4 | 3.265E-7 | 0.01 | -27472 | gtex_brain_ba24 |
| rs7515643 | 1 | 65866477 | LEPROT | ENSG00000213625.4 | 3.264E-7 | 0.01 | -26988 | gtex_brain_ba24 |
| rs12136950 | 1 | 65866516 | LEPROT | ENSG00000213625.4 | 3.265E-7 | 0.01 | -26949 | gtex_brain_ba24 |
| rs4592284 | 1 | 65867391 | LEPROT | ENSG00000213625.4 | 7.203E-7 | 0.01 | -26074 | gtex_brain_ba24 |
| rs1033997 | 1 | 65869163 | LEPROT | ENSG00000213625.4 | 1.747E-6 | 0.01 | -24302 | gtex_brain_ba24 |
| rs71058403 | 1 | 65834062 | LEPROT | ENSG00000213625.4 | 4.583E-6 | 0.01 | -59403 | gtex_brain_putamen_basal |
| rs9326063 | 1 | 65835911 | LEPROT | ENSG00000213625.4 | 4.013E-6 | 0.01 | -57554 | gtex_brain_putamen_basal |
| rs10158872 | 1 | 65836230 | LEPROT | ENSG00000213625.4 | 4.077E-6 | 0.01 | -57235 | gtex_brain_putamen_basal |
| rs10158873 | 1 | 65836244 | LEPROT | ENSG00000213625.4 | 6.313E-7 | 0.01 | -57221 | gtex_brain_putamen_basal |
| rs11208637 | 1 | 65836553 | LEPROT | ENSG00000213625.4 | 4.137E-6 | 0.01 | -56912 | gtex_brain_putamen_basal |
| rs8179492 | 1 | 65836908 | LEPROT | ENSG00000213625.4 | 4.077E-6 | 0.01 | -56557 | gtex_brain_putamen_basal |
| rs6588136 | 1 | 65837119 | LEPROT | ENSG00000213625.4 | 4.077E-6 | 0.01 | -56346 | gtex_brain_putamen_basal |
| rs6588137 | 1 | 65837282 | LEPROT | ENSG00000213625.4 | 6.313E-7 | 0.01 | -56183 | gtex_brain_putamen_basal |
| rs6693498 | 1 | 65837389 | LEPROT | ENSG00000213625.4 | 4.077E-6 | 0.01 | -56076 | gtex_brain_putamen_basal |
| rs7537733 | 1 | 65838229 | LEPROT | ENSG00000213625.4 | 4.077E-6 | 0.01 | -55236 | gtex_brain_putamen_basal |
| rs6588138 | 1 | 65838366 | LEPROT | ENSG00000213625.4 | 3.825E-6 | 0.01 | -55099 | gtex_brain_putamen_basal |
| rs6659883 | 1 | 65838618 | LEPROT | ENSG00000213625.4 | 3.395E-6 | 0.01 | -54847 | gtex_brain_putamen_basal |
| rs1591364 | 1 | 65842364 | LEPROT | ENSG00000213625.4 | 4.577E-7 | 0.01 | -51101 | gtex_brain_putamen_basal |
| rs716592 | 1 | 65843982 | LEPROT | ENSG00000213625.4 | 4.555E-7 | 0.01 | -49483 | gtex_brain_putamen_basal |
| rs3818449 | 1 | 65844963 | LEPROT | ENSG00000213625.4 | 2.426E-6 | 0.01 | -48502 | gtex_brain_putamen_basal |
| rs1334879 | 1 | 65846051 | LEPROT | ENSG00000213625.4 | 1.142E-6 | 0.01 | -47414 | gtex_brain_putamen_basal |
| rs1334878 | 1 | 65846124 | LEPROT | ENSG00000213625.4 | 1.166E-6 | 0.01 | -47341 | gtex_brain_putamen_basal |
| rs1334877 | 1 | 65846406 | LEPROT | ENSG00000213625.4 | 1.168E-6 | 0.01 | -47059 | gtex_brain_putamen_basal |
| rs11208640 | 1 | 65846996 | LEPROT | ENSG00000213625.4 | 1.432E-7 | 0.01 | -46469 | gtex_brain_putamen_basal |
| rs9728544 | 1 | 65847105 | LEPROT | ENSG00000213625.4 | 1.362E-7 | 0.01 | -46360 | gtex_brain_putamen_basal |
| rs9725541 | 1 | 65847186 | LEPROT | ENSG00000213625.4 | 9.63E-7 | 0.01 | -46279 | gtex_brain_putamen_basal |
| rs6697416 | 1 | 65848075 | LEPROT | ENSG00000213625.4 | 1.85E-6 | 0.01 | -45390 | gtex_brain_putamen_basal |
| rs11208642 | 1 | 65848622 | LEPROT | ENSG00000213625.4 | 1.845E-6 | 0.01 | -44843 | gtex_brain_putamen_basal |
| rs4593866 | 1 | 65850968 | LEPROT | ENSG00000213625.4 | 1.375E-7 | 0.01 | -42497 | gtex_brain_putamen_basal |
| rs10889546 | 1 | 65851759 | LEPROT | ENSG00000213625.4 | 1.107E-6 | 0.01 | -41706 | gtex_brain_putamen_basal |
| rs6700226 | 1 | 65852064 | LEPROT | ENSG00000213625.4 | 1.83E-6 | 0.01 | -41401 | gtex_brain_putamen_basal |
| rs11208643 | 1 | 65852310 | LEPROT | ENSG00000213625.4 | 4.056E-6 | 0.01 | -41155 | gtex_brain_putamen_basal |
| rs9436725 | 1 | 65852870 | LEPROT | ENSG00000213625.4 | 1.107E-6 | 0.01 | -40595 | gtex_brain_putamen_basal |
| rs9436726 | 1 | 65853196 | LEPROT | ENSG00000213625.4 | 1.107E-6 | 0.01 | -40269 | gtex_brain_putamen_basal |
| rs6588142 | 1 | 65854473 | LEPROT | ENSG00000213625.4 | 1.83E-6 | 0.01 | -38992 | gtex_brain_putamen_basal |
| rs6588143 | 1 | 65854556 | LEPROT | ENSG00000213625.4 | 1.83E-6 | 0.01 | -38909 | gtex_brain_putamen_basal |
| rs7545397 | 1 | 65856282 | LEPROT | ENSG00000213625.4 | 8.85E-7 | 0.01 | -37183 | gtex_brain_putamen_basal |
| rs7545720 | 1 | 65856443 | LEPROT | ENSG00000213625.4 | 7.62E-7 | 0.01 | -37022 | gtex_brain_putamen_basal |
| rs10789183 | 1 | 65856779 | LEPROT | ENSG00000213625.4 | 6.817E-7 | 0.01 | -36686 | gtex_brain_putamen_basal |
| rs4554758 | 1 | 65857403 | LEPROT | ENSG00000213625.4 | 2.329E-6 | 0.01 | -36062 | gtex_brain_putamen_basal |
| rs4575118 | 1 | 65857457 | LEPROT | ENSG00000213625.4 | 2.329E-6 | 0.01 | -36008 | gtex_brain_putamen_basal |
| rs4512686 | 1 | 65857483 | LEPROT | ENSG00000213625.4 | 2.329E-6 | 0.01 | -35982 | gtex_brain_putamen_basal |
| rs6588144 | 1 | 65858088 | LEPROT | ENSG00000213625.4 | 2.329E-6 | 0.01 | -35377 | gtex_brain_putamen_basal |
| rs4332387 | 1 | 65859108 | LEPROT | ENSG00000213625.4 | 8.534E-7 | 0.01 | -34357 | gtex_brain_putamen_basal |
| rs6588145 | 1 | 65859784 | LEPROT | ENSG00000213625.4 | 1.394E-7 | 0.01 | -33681 | gtex_brain_putamen_basal |
| rs4329545 | 1 | 65861060 | LEPROT | ENSG00000213625.4 | 6.205E-7 | 0.01 | -32405 | gtex_brain_putamen_basal |
| rs4916052 | 1 | 65865940 | LEPROT | ENSG00000213625.4 | 1.007E-6 | 0.01 | -27525 | gtex_brain_putamen_basal |
| rs4592284 | 1 | 65867391 | LEPROT | ENSG00000213625.4 | 1.568E-7 | 0.01 | -26074 | gtex_brain_putamen_basal |
| rs1033997 | 1 | 65869163 | LEPROT | ENSG00000213625.4 | 3.473E-6 | 0.01 | -24302 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
General gene expression (GTEx)
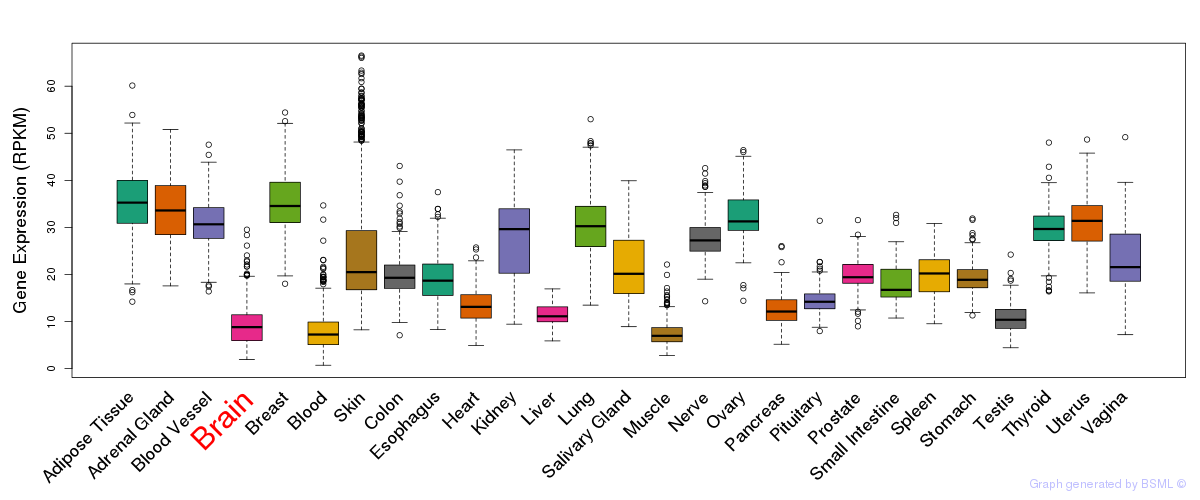
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003674 | molecular_function | ND | 9207021 | |
| GO:0005515 | protein binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008203 | cholesterol metabolic process | IEA | - | |
| GO:0008150 | biological_process | ND | 9207021 | |
| GO:0051346 | negative regulation of hydrolase activity | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | NAS | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA PROGENITOR UP | 39 | 25 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES DN | 145 | 91 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED UP | 78 | 48 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION UP | 119 | 66 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL DN | 86 | 59 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 UP | 276 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 UP | 341 | 197 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP | 164 | 122 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| INGRAM SHH TARGETS UP | 127 | 79 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| GOLUB ALL VS AML DN | 24 | 14 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 AND CD2 UP | 89 | 51 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 UP | 182 | 119 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 8 | 86 | 57 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD UP | 222 | 139 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A UP | 111 | 70 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE UP | 181 | 106 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 7 | 76 | 46 | All SZGR 2.0 genes in this pathway |
| RAGHAVACHARI PLATELET SPECIFIC GENES | 70 | 46 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS DN | 207 | 139 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |