Gene Page: PPP3CB
Summary ?
| GeneID | 5532 |
| Symbol | PPP3CB |
| Synonyms | CALNA2|CALNB|CNA2|PP2Bbeta |
| Description | protein phosphatase 3 catalytic subunit beta |
| Reference | MIM:114106|HGNC:HGNC:9315|Ensembl:ENSG00000107758|HPRD:00235|Vega:OTTHUMG00000018472 |
| Gene type | protein-coding |
| Map location | 10q22.2 |
| Pascal p-value | 0.624 |
| Sherlock p-value | 0.941 |
| Fetal beta | -0.587 |
| eGene | Putamen basal ganglia |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12357530 | 10 | 75928654 | PPP3CB | ENSG00000107758.11 | 9.796E-6 | 0.03 | -672872 | gtex_brain_putamen_basal |
| rs72351608 | 10 | 75928942 | PPP3CB | ENSG00000107758.11 | 9.796E-6 | 0.03 | -673160 | gtex_brain_putamen_basal |
| rs2395077 | 10 | 75932001 | PPP3CB | ENSG00000107758.11 | 9.796E-6 | 0.03 | -676219 | gtex_brain_putamen_basal |
| rs10824096 | 10 | 75937554 | PPP3CB | ENSG00000107758.11 | 9.796E-6 | 0.03 | -681772 | gtex_brain_putamen_basal |
| rs12359456 | 10 | 75937994 | PPP3CB | ENSG00000107758.11 | 9.796E-6 | 0.03 | -682212 | gtex_brain_putamen_basal |
| rs9665468 | 10 | 75938838 | PPP3CB | ENSG00000107758.11 | 9.796E-6 | 0.03 | -683056 | gtex_brain_putamen_basal |
| rs10740421 | 10 | 75941002 | PPP3CB | ENSG00000107758.11 | 5.646E-6 | 0.03 | -685220 | gtex_brain_putamen_basal |
| rs11000909 | 10 | 75944828 | PPP3CB | ENSG00000107758.11 | 9.796E-6 | 0.03 | -689046 | gtex_brain_putamen_basal |
| rs10824101 | 10 | 75947438 | PPP3CB | ENSG00000107758.11 | 9.796E-6 | 0.03 | -691656 | gtex_brain_putamen_basal |
| rs11000912 | 10 | 75948477 | PPP3CB | ENSG00000107758.11 | 9.796E-6 | 0.03 | -692695 | gtex_brain_putamen_basal |
| rs10824102 | 10 | 75948915 | PPP3CB | ENSG00000107758.11 | 9.796E-6 | 0.03 | -693133 | gtex_brain_putamen_basal |
| rs10824103 | 10 | 75949415 | PPP3CB | ENSG00000107758.11 | 9.796E-6 | 0.03 | -693633 | gtex_brain_putamen_basal |
| rs10824104 | 10 | 75952066 | PPP3CB | ENSG00000107758.11 | 9.796E-6 | 0.03 | -696284 | gtex_brain_putamen_basal |
| rs12220238 | 10 | 75952670 | PPP3CB | ENSG00000107758.11 | 9.796E-6 | 0.03 | -696888 | gtex_brain_putamen_basal |
| rs10824105 | 10 | 75952881 | PPP3CB | ENSG00000107758.11 | 9.796E-6 | 0.03 | -697099 | gtex_brain_putamen_basal |
| rs7073227 | 10 | 75953047 | PPP3CB | ENSG00000107758.11 | 9.796E-6 | 0.03 | -697265 | gtex_brain_putamen_basal |
| rs7073432 | 10 | 75953350 | PPP3CB | ENSG00000107758.11 | 9.796E-6 | 0.03 | -697568 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
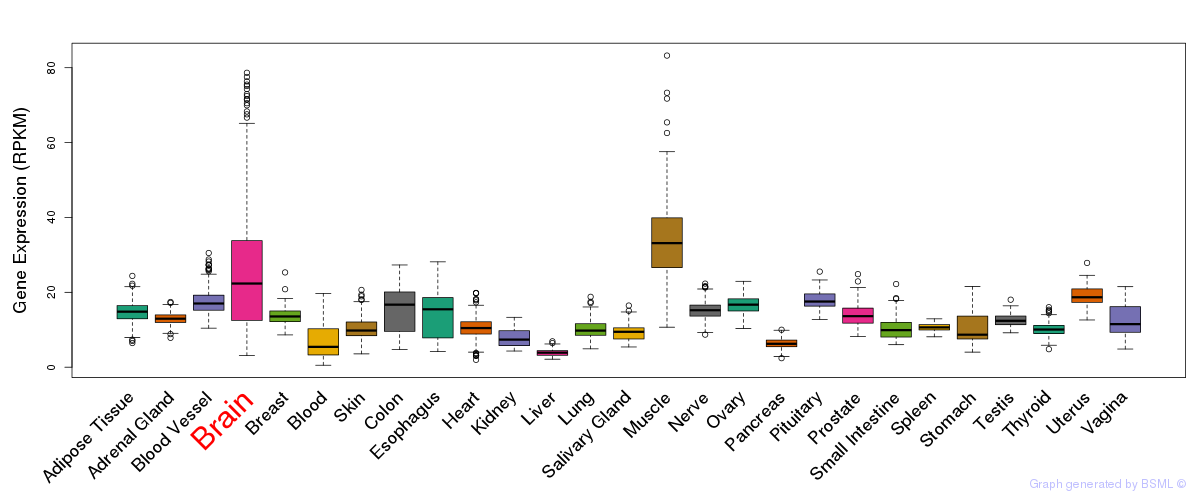
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DLSTP | 0.92 | 0.93 |
| RQCD1 | 0.91 | 0.92 |
| C20orf4 | 0.91 | 0.92 |
| ACTR1A | 0.90 | 0.90 |
| EFTUD2 | 0.90 | 0.90 |
| WDR40A | 0.90 | 0.89 |
| EDC3 | 0.90 | 0.91 |
| GLE1 | 0.90 | 0.90 |
| KIAA1191 | 0.90 | 0.89 |
| SEC31A | 0.90 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.78 | -0.85 |
| MT-CO2 | -0.77 | -0.86 |
| AF347015.27 | -0.76 | -0.85 |
| MT-CYB | -0.75 | -0.84 |
| AF347015.8 | -0.74 | -0.85 |
| AF347015.33 | -0.74 | -0.81 |
| AF347015.15 | -0.71 | -0.82 |
| AF347015.21 | -0.71 | -0.87 |
| AF347015.2 | -0.70 | -0.81 |
| C5orf53 | -0.70 | -0.72 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG AXON GUIDANCE | 129 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG VEGF SIGNALING PATHWAY | 76 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG NATURAL KILLER CELL MEDIATED CYTOTOXICITY | 137 | 92 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| KEGG B CELL RECEPTOR SIGNALING PATHWAY | 75 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM POTENTIATION | 70 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| KEGG AMYOTROPHIC LATERAL SCLEROSIS ALS | 53 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BCR PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HDAC PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CALCINEURIN PATHWAY | 21 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NDKDYNAMIN PATHWAY | 21 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FCER1 PATHWAY | 41 | 30 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FMLP PATHWAY | 39 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA VIP PATHWAY | 29 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NFAT PATHWAY | 56 | 45 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NOS1 PATHWAY | 24 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PGC1A PATHWAY | 26 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MEF2D PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GPCR PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TCR PATHWAY | 49 | 37 | All SZGR 2.0 genes in this pathway |
| SIG BCR SIGNALING PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| PID BCR 5PATHWAY | 65 | 50 | All SZGR 2.0 genes in this pathway |
| PID IL12 2PATHWAY | 63 | 54 | All SZGR 2.0 genes in this pathway |
| PID NFAT 3PATHWAY | 54 | 47 | All SZGR 2.0 genes in this pathway |
| PID ERBB2 ERBB3 PATHWAY | 44 | 35 | All SZGR 2.0 genes in this pathway |
| PID TCR CALCIUM PATHWAY | 29 | 23 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR DOWNSTREAM PATHWAY | 65 | 56 | All SZGR 2.0 genes in this pathway |
| PID IL12 STAT4 PATHWAY | 33 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME OPIOID SIGNALLING | 78 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME DARPP 32 EVENTS | 25 | 20 | All SZGR 2.0 genes in this pathway |
| GRABARCZYK BCL11B TARGETS DN | 57 | 35 | All SZGR 2.0 genes in this pathway |
| RODRIGUES DCC TARGETS DN | 121 | 84 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS UP | 135 | 82 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| KIM GERMINAL CENTER T HELPER UP | 66 | 42 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| ZHAN VARIABLE EARLY DIFFERENTIATION GENES DN | 30 | 19 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A | 182 | 108 | All SZGR 2.0 genes in this pathway |
| VANOEVELEN MYOGENESIS SIN3A TARGETS | 220 | 133 | All SZGR 2.0 genes in this pathway |