Gene Page: CHST12
Summary ?
| GeneID | 55501 |
| Symbol | CHST12 |
| Synonyms | C4S-2|C4ST-2|C4ST2 |
| Description | carbohydrate (chondroitin 4) sulfotransferase 12 |
| Reference | MIM:610129|HGNC:HGNC:17423|Ensembl:ENSG00000136213|HPRD:13056| |
| Gene type | protein-coding |
| Map location | 7p22 |
| Pascal p-value | 0.002 |
| Sherlock p-value | 0.896 |
| Fetal beta | 0.118 |
| DMG | 2 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 2 |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg12619262 | 7 | 2480493 | CHST12 | 2.85E-6 | 0.016 | 0.024 | DMG:Montano_2016 |
| cg24743283 | 7 | 2884365 | CHST12 | 4.572E-4 | 5.744 | DMG:vanEijk_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | CHST12 | 55501 | 2.377E-4 | trans | ||
| rs6797307 | chr3 | 8601563 | CHST12 | 55501 | 0.12 | trans | ||
| rs6997759 | chr8 | 1184045 | CHST12 | 55501 | 0.19 | trans | ||
| rs2766987 | chr9 | 113667956 | CHST12 | 55501 | 0.13 | trans | ||
| rs16955618 | chr15 | 29937543 | CHST12 | 55501 | 7.045E-6 | trans |
Section II. Transcriptome annotation
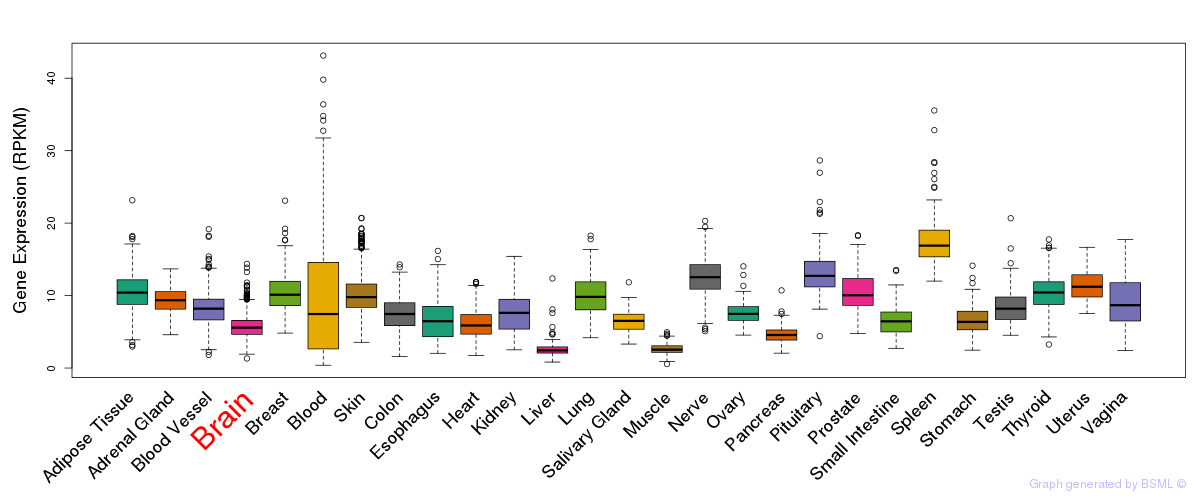
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C3orf57 | 0.86 | 0.81 |
| AC112491.3 | 0.86 | 0.76 |
| BTBD11 | 0.86 | 0.81 |
| GRIA4 | 0.85 | 0.82 |
| C1orf201 | 0.83 | 0.79 |
| STS | 0.83 | 0.78 |
| TPD52 | 0.82 | 0.83 |
| KCNC2 | 0.81 | 0.79 |
| MAP7D2 | 0.80 | 0.82 |
| OLFM3 | 0.80 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SIGIRR | -0.41 | -0.50 |
| NME4 | -0.40 | -0.53 |
| CDC42EP4 | -0.39 | -0.45 |
| WDR86 | -0.38 | -0.40 |
| EFEMP2 | -0.38 | -0.43 |
| SMTN | -0.37 | -0.45 |
| SEMA4B | -0.36 | -0.34 |
| RPL23A | -0.35 | -0.41 |
| GPR125 | -0.35 | -0.32 |
| C6orf48 | -0.35 | -0.29 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCOSAMINOGLYCAN BIOSYNTHESIS CHONDROITIN SULFATE | 22 | 16 | All SZGR 2.0 genes in this pathway |
| KEGG SULFUR METABOLISM | 13 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | 21 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | 49 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOSAMINOGLYCAN METABOLISM | 111 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 7P22 AMPLICON | 38 | 27 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 9 | 35 | 26 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS DN | 213 | 127 | All SZGR 2.0 genes in this pathway |
| ACOSTA PROLIFERATION INDEPENDENT MYC TARGETS DN | 116 | 74 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |