Gene Page: ANKRD10
Summary ?
| GeneID | 55608 |
| Symbol | ANKRD10 |
| Synonyms | - |
| Description | ankyrin repeat domain 10 |
| Reference | HGNC:HGNC:20265|Ensembl:ENSG00000088448|HPRD:12455|Vega:OTTHUMG00000017349 |
| Gene type | protein-coding |
| Map location | 13q34 |
| Pascal p-value | 0.003 |
| Sherlock p-value | 0.325 |
| Fetal beta | 1.06 |
| DMG | 2 (# studies) |
| eGene | Cortex Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04467034 | 13 | 111567140 | ANKRD10 | -0.023 | 0.45 | DMG:Nishioka_2013 | |
| cg06825833 | 13 | 111566802 | ANKRD10 | 1E-7 | -0.011 | 2.22E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs336544 | chr3 | 161060557 | ANKRD10 | 55608 | 0.19 | trans |
Section II. Transcriptome annotation
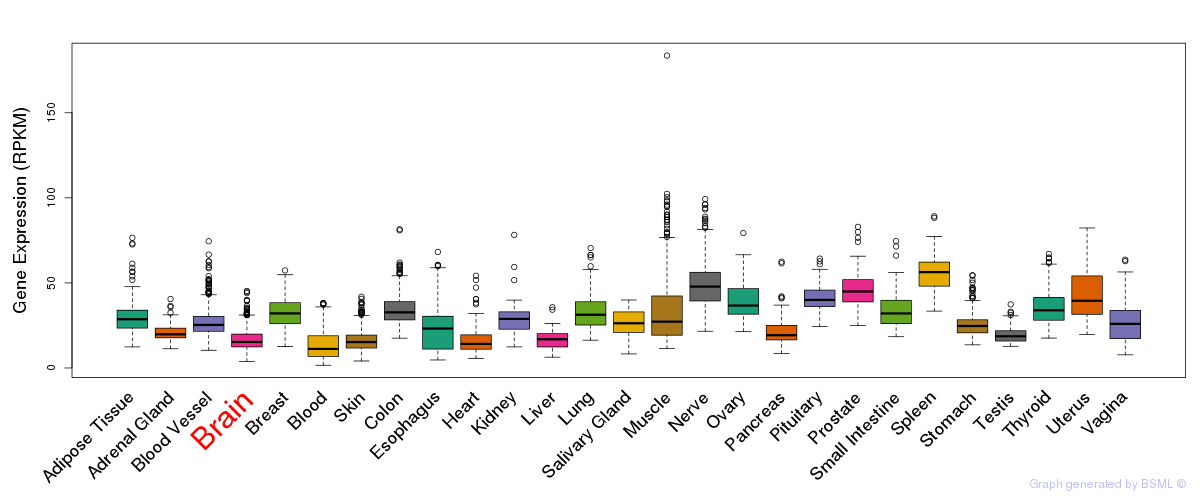
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FAM117B | 0.98 | 0.88 |
| SLCO5A1 | 0.96 | 0.88 |
| KLF11 | 0.96 | 0.85 |
| ZSCAN2 | 0.95 | 0.90 |
| KDM5B | 0.95 | 0.87 |
| DCX | 0.95 | 0.90 |
| IGSF3 | 0.95 | 0.91 |
| EIF2C4 | 0.95 | 0.87 |
| CEP170 | 0.95 | 0.87 |
| PRRC1 | 0.95 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LHPP | -0.68 | -0.64 |
| C5orf53 | -0.68 | -0.78 |
| FBXO2 | -0.68 | -0.69 |
| HLA-F | -0.68 | -0.79 |
| ALDOC | -0.67 | -0.72 |
| AIFM3 | -0.67 | -0.76 |
| LDHD | -0.66 | -0.67 |
| PTH1R | -0.66 | -0.73 |
| SLC16A11 | -0.66 | -0.67 |
| CLU | -0.66 | -0.71 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA DN | 77 | 52 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA COPY NUMBER DN | 54 | 37 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER EXPRESSION BY COPY NUMBER | 100 | 62 | All SZGR 2.0 genes in this pathway |
| ZUCCHI METASTASIS UP | 43 | 24 | All SZGR 2.0 genes in this pathway |
| KUROKAWA LIVER CANCER CHEMOTHERAPY UP | 12 | 8 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT DN | 129 | 86 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR | 293 | 193 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| CAMPS COLON CANCER COPY NUMBER UP | 92 | 45 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| CHANG CYCLING GENES | 148 | 83 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| LEE RECENT THYMIC EMIGRANT | 227 | 128 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G1 S | 147 | 76 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BAKKER FOXO3 TARGETS DN | 187 | 109 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL DN | 146 | 88 | All SZGR 2.0 genes in this pathway |