Gene Page: PRKAR1A
Summary ?
| GeneID | 5573 |
| Symbol | PRKAR1A |
| Synonyms | ACRDYS1|ADOHR|CAR|CNC|CNC1|PKR1|PPNAD1|PRKAR1|TSE1 |
| Description | protein kinase cAMP-dependent type I regulatory subunit alpha |
| Reference | MIM:188830|HGNC:HGNC:9388|Ensembl:ENSG00000108946|HPRD:01786|Vega:OTTHUMG00000180128 |
| Gene type | protein-coding |
| Map location | 17q24.2 |
| Pascal p-value | 0.163 |
| Sherlock p-value | 0.299 |
| Fetal beta | -1.349 |
| Support | CANABINOID DOPAMINE INTRACELLULAR SIGNAL TRANSDUCTION SEROTONIN G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
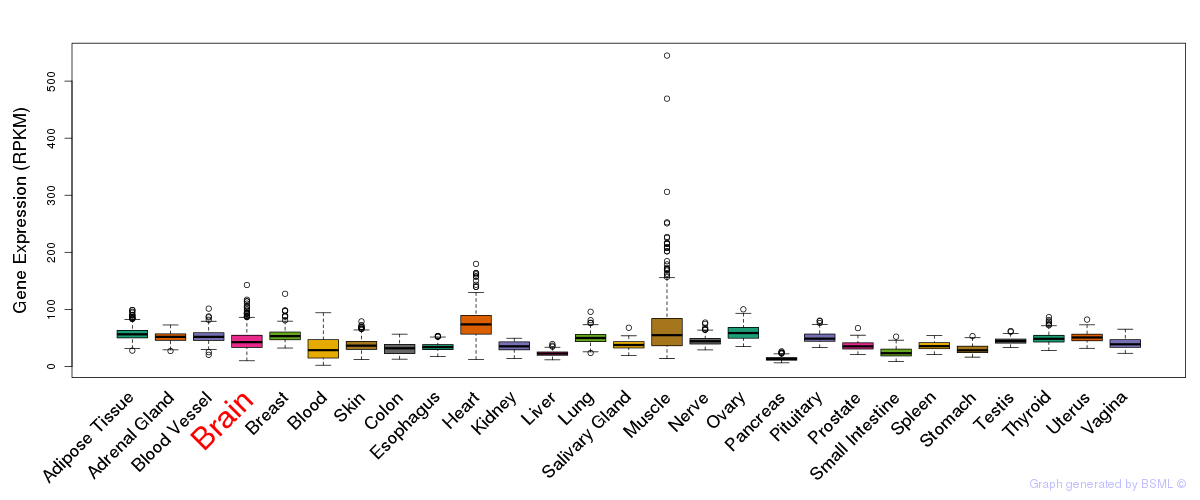
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ADRM1 | 0.91 | 0.89 |
| POLR2E | 0.90 | 0.89 |
| GPS1 | 0.90 | 0.89 |
| TOMM40 | 0.90 | 0.90 |
| THAP4 | 0.89 | 0.87 |
| TRAP1 | 0.89 | 0.88 |
| FAM69B | 0.89 | 0.90 |
| IDH3G | 0.88 | 0.86 |
| MRPS2 | 0.88 | 0.88 |
| TCF25 | 0.88 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.73 | -0.73 |
| AF347015.8 | -0.71 | -0.73 |
| AF347015.33 | -0.71 | -0.74 |
| MT-CO2 | -0.71 | -0.71 |
| AF347015.27 | -0.71 | -0.75 |
| MT-CYB | -0.70 | -0.73 |
| AF347015.21 | -0.69 | -0.72 |
| AF347015.2 | -0.68 | -0.71 |
| AF347015.15 | -0.67 | -0.71 |
| AF347015.26 | -0.64 | -0.68 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKAP1 | AKAP | AKAP121 | AKAP149 | AKAP84 | D-AKAP1 | MGC1807 | PRKA1 | SAKAP84 | A kinase (PRKA) anchor protein 1 | - | HPRD | 10764601 |12634056|12634056 |
| AKAP1 | AKAP | AKAP121 | AKAP149 | AKAP84 | D-AKAP1 | MGC1807 | PRKA1 | SAKAP84 | A kinase (PRKA) anchor protein 1 | - | HPRD,BioGRID | 9065479 |12634056|10764601 |12634056|12634056 |
| AKAP10 | D-AKAP2 | MGC9414 | PRKA10 | A kinase (PRKA) anchor protein 10 | - | HPRD,BioGRID | 9326583 |
| AKAP11 | AKAP220 | DKFZp781I12161 | FLJ11304 | KIAA0629 | PRKA11 | A kinase (PRKA) anchor protein 11 | - | HPRD,BioGRID | 10864471 |
| AKAP4 | AKAP82 | FSC1 | HI | hAKAP82 | p82 | A kinase (PRKA) anchor protein 4 | - | HPRD,BioGRID | 9852104 |
| AKAP7 | AKAP18 | A kinase (PRKA) anchor protein 7 | - | HPRD | 12804576 |
| ARFGEF1 | ARFGEP1 | BIG1 | D730028O18Rik | DKFZP434L057 | P200 | ADP-ribosylation factor guanine nucleotide-exchange factor 1(brefeldin A-inhibited) | Affinity Capture-Western | BioGRID | 12571360 |
| ARFGEF2 | BIG2 | FLJ23723 | dJ1164I10.1 | ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12571360 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD,BioGRID | 9050991 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD,BioGRID | 9050991 |
| MCRS1 | ICP22BP | INO80Q | MCRS2 | MSP58 | P78 | microspherule protein 1 | Affinity Capture-MS | BioGRID | 17353931 |
| MGC13057 | - | hypothetical protein MGC13057 | Two-hybrid | BioGRID | 16189514 |
| MYO7A | DFNA11 | DFNB2 | MYOVIIA | MYU7A | NSRD2 | USH1B | myosin VIIA | Affinity Capture-Western Two-hybrid | BioGRID | 10889203 |
| NUAK1 | ARK5 | KIAA0537 | NUAK family, SNF1-like kinase, 1 | Affinity Capture-MS | BioGRID | 17353931 |
| PLEKHF2 | FLJ13187 | PHAFIN2 | ZFYVE18 | pleckstrin homology domain containing, family F (with FYVE domain) member 2 | Two-hybrid | BioGRID | 16189514 |
| PRKACA | MGC102831 | MGC48865 | PKACA | protein kinase, cAMP-dependent, catalytic, alpha | Affinity Capture-MS | BioGRID | 17353931 |
| PRKAR1A | CAR | CNC | CNC1 | DKFZp779L0468 | MGC17251 | PKR1 | PPNAD1 | PRKAR1 | TSE1 | protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) | Affinity Capture-Western Two-hybrid | BioGRID | 12634056 |
| PRKAR1B | PRKAR1 | protein kinase, cAMP-dependent, regulatory, type I, beta | - | HPRD | 12634056 |
| PRKAR1B | PRKAR1 | protein kinase, cAMP-dependent, regulatory, type I, beta | - | HPRD,BioGRID | 8407966 |12634056 |
| PRKX | PKX1 | protein kinase, X-linked | Reconstituted Complex | BioGRID | 10026146 |
| PYCARD | ASC | CARD5 | MGC10332 | TMS | TMS-1 | TMS1 | PYD and CARD domain containing | Two-hybrid | BioGRID | 16189514 |
| TUSC4 | NPR2L | NPRL2 | tumor suppressor candidate 4 | Affinity Capture-MS | BioGRID | 17353931 |
| UBE2M | UBC-RS2 | UBC12 | hUbc12 | ubiquitin-conjugating enzyme E2M (UBC12 homolog, yeast) | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NO1 PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CSK PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AGPCR PATHWAY | 13 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CFTR PATHWAY | 12 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GATA3 PATHWAY | 16 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MPR PATHWAY | 34 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MCALPAIN PATHWAY | 25 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PPARA PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IGF1R PATHWAY | 23 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA VIP PATHWAY | 29 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NFAT PATHWAY | 56 | 45 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NOS1 PATHWAY | 24 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PLCE PATHWAY | 12 | 10 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CHREBP2 PATHWAY | 42 | 35 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BAD PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CK1 PATHWAY | 17 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DREAM PATHWAY | 14 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GPCR PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SHH PATHWAY | 16 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA STATHMIN PATHWAY | 19 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CREB PATHWAY | 27 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CARM1 PATHWAY | 13 | 11 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN A4B1 PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME DAG AND IP3 SIGNALING | 32 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2 | 101 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER | 109 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE | 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRATION OF ENERGY METABOLISM | 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME OPIOID SIGNALLING | 78 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME CA DEPENDENT EVENTS | 30 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME DARPP 32 EVENTS | 25 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME PLC BETA MEDIATED EVENTS | 43 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | 18 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | 34 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF | 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | 95 | 76 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | 43 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION | 93 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | 100 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPASE C MEDIATED CASCADE | 54 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME AQUAPORIN MEDIATED TRANSPORT | 51 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | 132 | 101 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR | 112 | 80 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP | 372 | 227 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D UP | 175 | 108 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP | 185 | 126 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 UP | 64 | 46 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| XU AKT1 TARGETS 6HR | 27 | 18 | All SZGR 2.0 genes in this pathway |
| SIMBULAN UV RESPONSE IMMORTALIZED DN | 31 | 26 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| SAKAI TUMOR INFILTRATING MONOCYTES DN | 81 | 51 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| PARK HSC VS MULTIPOTENT PROGENITORS UP | 19 | 14 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR UP | 151 | 100 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| VISALA RESPONSE TO HEAT SHOCK AND AGING UP | 15 | 10 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| MMS MOUSE LYMPH HIGH 4HRS UP | 36 | 20 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC UP | 123 | 75 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE UP | 126 | 92 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOZOA | 114 | 77 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP | 198 | 128 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G3 UP | 188 | 121 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS UP | 91 | 59 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN UP | 90 | 58 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |