Gene Page: MCTP2
Summary ?
| GeneID | 55784 |
| Symbol | MCTP2 |
| Synonyms | - |
| Description | multiple C2 domains, transmembrane 2 |
| Reference | MIM:616297|HGNC:HGNC:25636|Ensembl:ENSG00000140563|HPRD:07738|Vega:OTTHUMG00000171751 |
| Gene type | protein-coding |
| Map location | 15q26.2 |
| Pascal p-value | 0.918 |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
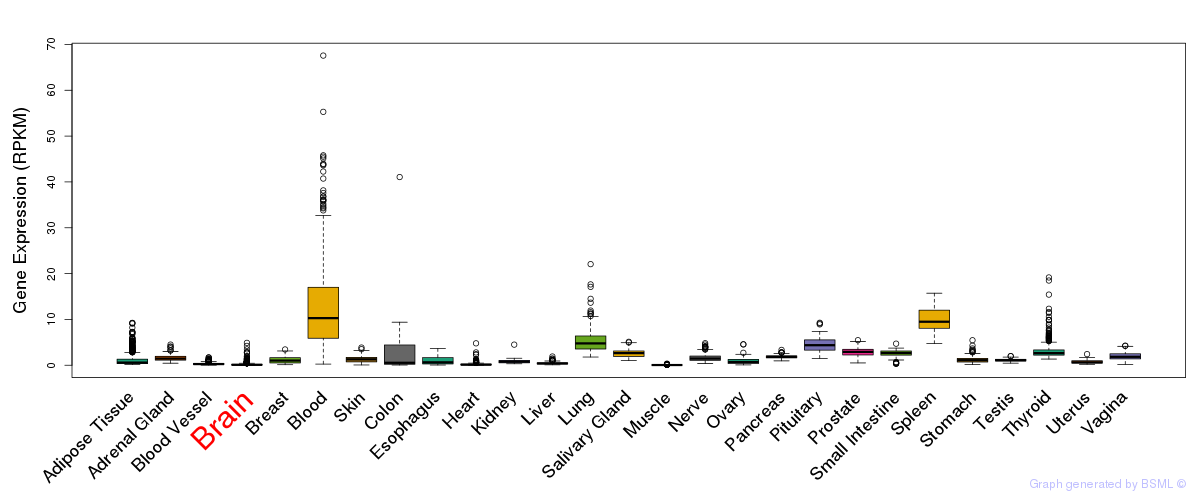
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PREPL | 0.92 | 0.93 |
| CHM | 0.91 | 0.91 |
| LMBRD2 | 0.91 | 0.91 |
| WASL | 0.91 | 0.91 |
| CACNB4 | 0.91 | 0.91 |
| EFHA2 | 0.90 | 0.91 |
| NCOA7 | 0.90 | 0.89 |
| DPP8 | 0.90 | 0.89 |
| NDFIP2 | 0.90 | 0.87 |
| SH3BGRL2 | 0.89 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EFEMP2 | -0.48 | -0.55 |
| RAB34 | -0.46 | -0.54 |
| NUDT8 | -0.45 | -0.52 |
| AL022328.1 | -0.44 | -0.58 |
| BCL7C | -0.43 | -0.47 |
| AC006276.2 | -0.42 | -0.41 |
| C16orf79 | -0.42 | -0.50 |
| IRF7 | -0.41 | -0.56 |
| RAMP2 | -0.41 | -0.46 |
| C1orf61 | -0.41 | -0.58 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| CASORELLI APL SECONDARY VS DE NOVO UP | 39 | 25 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR DN | 214 | 133 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA DN | 284 | 156 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P4 | 100 | 62 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE DN | 204 | 114 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| VANOEVELEN MYOGENESIS SIN3A TARGETS | 220 | 133 | All SZGR 2.0 genes in this pathway |
| GHANDHI BYSTANDER IRRADIATION UP | 86 | 54 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |