Gene Page: PAG1
Summary ?
| GeneID | 55824 |
| Symbol | PAG1 |
| Synonyms | CBP|PAG |
| Description | phosphoprotein membrane anchor with glycosphingolipid microdomains 1 |
| Reference | MIM:605767|HGNC:HGNC:30043|Ensembl:ENSG00000076641|HPRD:05772|HPRD:16944|Vega:OTTHUMG00000164592 |
| Gene type | protein-coding |
| Map location | 8q21.13 |
| Pascal p-value | 0.159 |
| Sherlock p-value | 0.132 |
| Fetal beta | 1.061 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| DNM:Xu_2012 | Whole Exome Sequencing analysis | De novo mutations of 4 genes were identified by exome sequencing of 795 samples in this study | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PAG1 | chr8 | 81905378 | C | T | NM_018440 | p.29V>I | missense | Schizophrenia | DNM:Xu_2012 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg07618730 | 8 | 82023929 | PAG1 | 5.169E-4 | -0.324 | 0.047 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12405921 | chr1 | 206695730 | PAG1 | 55824 | 0.05 | trans | ||
| rs16829545 | chr2 | 151977407 | PAG1 | 55824 | 1.594E-12 | trans | ||
| rs3845734 | chr2 | 171125572 | PAG1 | 55824 | 0.02 | trans | ||
| rs7584986 | chr2 | 184111432 | PAG1 | 55824 | 1.362E-4 | trans | ||
| rs9461864 | chr6 | 33481468 | PAG1 | 55824 | 0.19 | trans | ||
| rs3118341 | chr9 | 25185518 | PAG1 | 55824 | 0.14 | trans | ||
| rs3765543 | chr9 | 134458323 | PAG1 | 55824 | 0.09 | trans | ||
| rs2393316 | chr10 | 59333070 | PAG1 | 55824 | 0.01 | trans | ||
| rs1526959 | chr12 | 79753789 | PAG1 | 55824 | 0.16 | trans | ||
| rs16955618 | chr15 | 29937543 | PAG1 | 55824 | 1.133E-14 | trans | ||
| rs1041786 | chr21 | 22617710 | PAG1 | 55824 | 0 | trans |
Section II. Transcriptome annotation
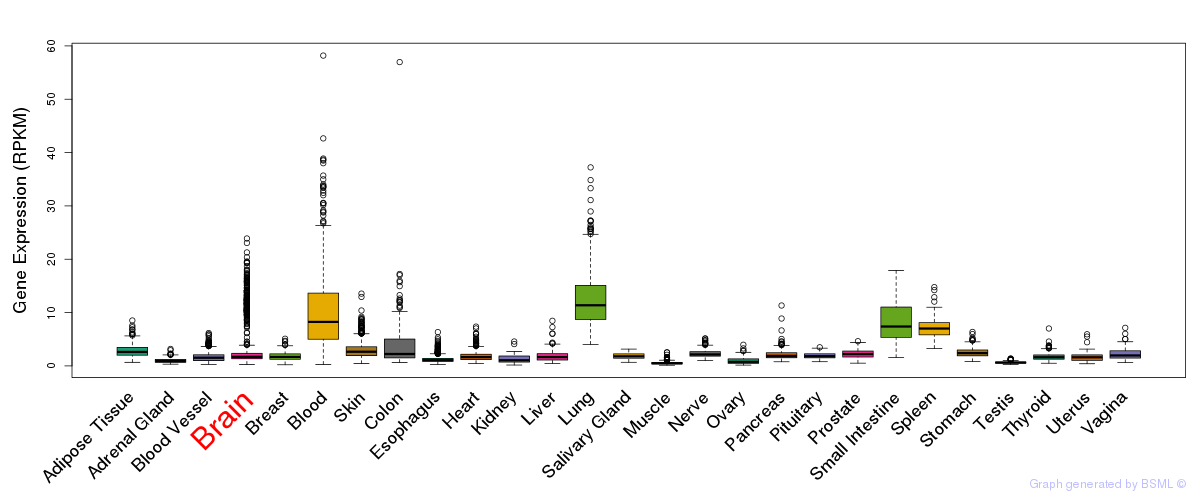
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | - | HPRD,BioGRID | 9334312 |
| CSK | MGC117393 | c-src tyrosine kinase | - | HPRD,BioGRID | 10790433 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD,BioGRID | 10790433 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD,BioGRID | 10790433 |
| LCK | YT16 | p56lck | pp58lck | lymphocyte-specific protein tyrosine kinase | - | HPRD,BioGRID | 10790433 |
| LCP2 | SLP-76 | SLP76 | lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) | - | HPRD,BioGRID | 10790433 |
| LYN | FLJ26625 | JTK8 | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | - | HPRD,BioGRID | 10790433 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | - | HPRD | 10790433 |
| PTPN6 | HCP | HCPH | HPTP1C | PTP-1C | SH-PTP1 | SHP-1 | SHP-1L | SHP1 | protein tyrosine phosphatase, non-receptor type 6 | - | HPRD,BioGRID | 10790433 |
| RASA1 | CM-AVM | CMAVM | DKFZp434N071 | GAP | PKWS | RASA | RASGAP | p120GAP | p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 | - | HPRD | 10790433 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD,BioGRID | 10790433 |
| SLC9A3R1 | EBP50 | NHERF | NHERF1 | NPHLOP2 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 | - | HPRD,BioGRID | 11684085 |
| SYK | DKFZp313N1010 | FLJ25043 | FLJ37489 | spleen tyrosine kinase | - | HPRD,BioGRID | 10790433 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | - | HPRD,BioGRID | 10790433 |
| ZAP70 | FLJ17670 | FLJ17679 | SRK | STD | TZK | ZAP-70 | zeta-chain (TCR) associated protein kinase 70kDa | - | HPRD,BioGRID | 10790433 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID BCR 5PATHWAY | 65 | 50 | All SZGR 2.0 genes in this pathway |
| PID TCR PATHWAY | 66 | 51 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR PATHWAY | 53 | 42 | All SZGR 2.0 genes in this pathway |
| PID SHP2 PATHWAY | 58 | 46 | All SZGR 2.0 genes in this pathway |
| PID CXCR4 PATHWAY | 102 | 78 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER | 109 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME TCR SIGNALING | 54 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME GAB1 SIGNALOSOME | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | 16 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| GRABARCZYK BCL11B TARGETS DN | 57 | 35 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA UP | 16 | 7 | All SZGR 2.0 genes in this pathway |
| FERREIRA EWINGS SARCOMA UNSTABLE VS STABLE UP | 167 | 92 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP | 223 | 140 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q12 Q22 AMPLICON | 132 | 82 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 2 | 114 | 76 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| HU GENOTOXIN ACTION DIRECT VS INDIRECT 24HR | 55 | 38 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS UP | 113 | 71 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS DN | 314 | 188 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| LEE DIFFERENTIATING T LYMPHOCYTE | 200 | 115 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 DN | 170 | 105 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION UP | 178 | 108 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS POLYSOMY7 DN | 24 | 17 | All SZGR 2.0 genes in this pathway |
| HOELZEL NF1 TARGETS UP | 139 | 93 | All SZGR 2.0 genes in this pathway |
| GABRIELY MIR21 TARGETS | 289 | 187 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |