Gene Page: C20orf24
Summary ?
| GeneID | 55969 |
| Symbol | C20orf24 |
| Synonyms | PNAS-11|RIP5 |
| Description | chromosome 20 open reading frame 24 |
| Reference | HGNC:HGNC:15870|Ensembl:ENSG00000101084|HPRD:12761|Vega:OTTHUMG00000032384 |
| Gene type | protein-coding |
| Map location | 20q11.23 |
| Pascal p-value | 0.624 |
| Sherlock p-value | 0.114 |
| Fetal beta | -0.364 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 6 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04759756 | 20 | 35273933 | C20orf24 | 3.48E-6 | 3.804 | DMG:vanEijk_2014 | |
| cg04759756 | 20 | 35273933 | C20orf24 | 1.79E-6 | 3.481 | DMG:vanEijk_2014 | |
| cg04759756 | 20 | 35273933 | C20orf24 | 1.66E-5 | 3.194 | DMG:vanEijk_2014 | |
| cg21671476 | 20 | 35169609 | C20orf24 | 5.382E-4 | -3.896 | DMG:vanEijk_2014 | |
| cg21671476 | 20 | 35169609 | C20orf24 | 8.014E-4 | -4.197 | DMG:vanEijk_2014 | |
| cg21671476 | 20 | 35169609 | C20orf24 | 6.3E-5 | -4.427 | DMG:vanEijk_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6902183 | chr6 | 121963786 | C20orf24 | 55969 | 0.16 | trans |
Section II. Transcriptome annotation
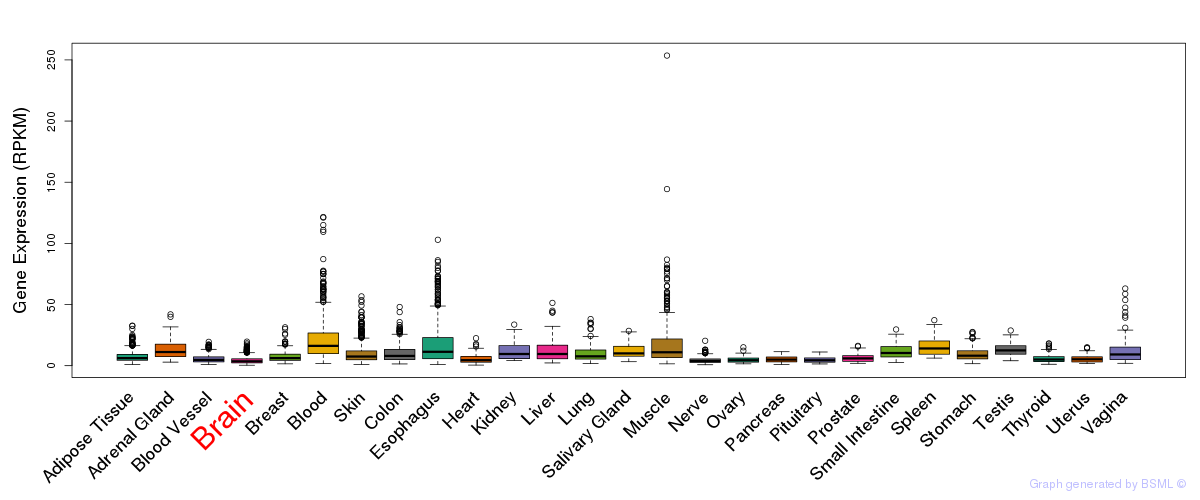
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABCD3 | ABC43 | PMP70 | PXMP1 | ATP-binding cassette, sub-family D (ALD), member 3 | Affinity Capture-MS | BioGRID | 17353931 |
| AIFM1 | AIF | MGC111425 | PDCD8 | apoptosis-inducing factor, mitochondrion-associated, 1 | Affinity Capture-MS | BioGRID | 17353931 |
| ATAD3A | FLJ10709 | ATPase family, AAA domain containing 3A | Affinity Capture-MS | BioGRID | 17353931 |
| ATAD3B | AAA-TOB3 | KIAA1273 | TOB3 | ATPase family, AAA domain containing 3B | Affinity Capture-MS | BioGRID | 17353931 |
| CLPX | - | ClpX caseinolytic peptidase X homolog (E. coli) | Affinity Capture-MS | BioGRID | 17353931 |
| DDX3X | DBX | DDX14 | DDX3 | HLP2 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked | Affinity Capture-MS | BioGRID | 17353931 |
| DYNC1H1 | DHC1 | DHC1a | DKFZp686P2245 | DNCH1 | DNCL | DNECL | DYHC | Dnchc1 | HL-3 | KIAA0325 | p22 | dynein, cytoplasmic 1, heavy chain 1 | Affinity Capture-MS | BioGRID | 17353931 |
| FANCI | FLJ10719 | KIAA1794 | Fanconi anemia, complementation group I | Affinity Capture-MS | BioGRID | 17353931 |
| FAR1 | DKFZp686A0370 | DKFZp686P18247 | FLJ22728 | FLJ33561 | MLSTD2 | fatty acyl CoA reductase 1 | Affinity Capture-MS | BioGRID | 17353931 |
| GCN1L1 | GCN1 | GCN1L | KIAA0219 | GCN1 general control of amino-acid synthesis 1-like 1 (yeast) | Affinity Capture-MS | BioGRID | 17353931 |
| IPO11 | RanBP11 | importin 11 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMD3 | P58 | RPN3 | S3 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 | Affinity Capture-MS | BioGRID | 17353931 |
| PTPLAD1 | B-IND1 | FLJ90376 | HSPC121 | protein tyrosine phosphatase-like A domain containing 1 | Affinity Capture-MS | BioGRID | 17353931 |
| RFXANK | ANKRA1 | BLS | F14150_1 | MGC138628 | RFX-B | regulatory factor X-associated ankyrin-containing protein | Affinity Capture-MS | BioGRID | 17353931 |
| SLC25A12 | ARALAR | ARALAR1 | solute carrier family 25 (mitochondrial carrier, Aralar), member 12 | Affinity Capture-MS | BioGRID | 17353931 |
| SPTLC1 | HSAN | HSAN1 | HSN1 | LBC1 | LCB1 | MGC14645 | SPT1 | SPTI | serine palmitoyltransferase, long chain base subunit 1 | Affinity Capture-MS | BioGRID | 17353931 |
| SSR3 | TRAPG | signal sequence receptor, gamma (translocon-associated protein gamma) | Affinity Capture-MS | BioGRID | 17353931 |
| SSR4 | TRAPD | signal sequence receptor, delta (translocon-associated protein delta) | Affinity Capture-MS | BioGRID | 17353931 |
| SURF4 | ERV29 | FLJ22993 | MGC102753 | surfeit 4 | Affinity Capture-MS | BioGRID | 17353931 |
| TNPO3 | IPO12 | MTR10A | TRN-SR | TRN-SR2 | TRNSR | transportin 3 | Affinity Capture-MS | BioGRID | 17353931 |
| XPO7 | KIAA0745 | RANBP16 | exportin 7 | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP | 285 | 181 | All SZGR 2.0 genes in this pathway |
| SOTIRIOU BREAST CANCER GRADE 1 VS 3 UP | 151 | 84 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER UP | 142 | 96 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW UP | 162 | 104 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH UP | 147 | 101 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA UP | 146 | 104 | All SZGR 2.0 genes in this pathway |
| CHOI ATL STAGE PREDICTOR | 44 | 24 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| GOLDRATH HOMEOSTATIC PROLIFERATION | 171 | 102 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C1 | 72 | 45 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |