Gene Page: NCEH1
Summary ?
| GeneID | 57552 |
| Symbol | NCEH1 |
| Synonyms | AADACL1|NCEH |
| Description | neutral cholesterol ester hydrolase 1 |
| Reference | MIM:613234|HGNC:HGNC:29260|Ensembl:ENSG00000144959|HPRD:07489|Vega:OTTHUMG00000156872 |
| Gene type | protein-coding |
| Map location | 3q26.31 |
| Pascal p-value | 0.032 |
| Sherlock p-value | 0.379 |
| DMG | 1 (# studies) |
| eGene | Cerebellum Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01232492 | 3 | 172429000 | NCEH1 | 8.03E-10 | -0.012 | 1.04E-6 | DMG:Jaffe_2016 |
| cg08037420 | 3 | 172428855 | NCEH1 | 3.53E-9 | -0.02 | 2.29E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs13023481 | chr2 | 213839772 | NCEH1 | 57552 | 0.19 | trans | ||
| rs10994209 | chr10 | 61877705 | NCEH1 | 57552 | 0.09 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
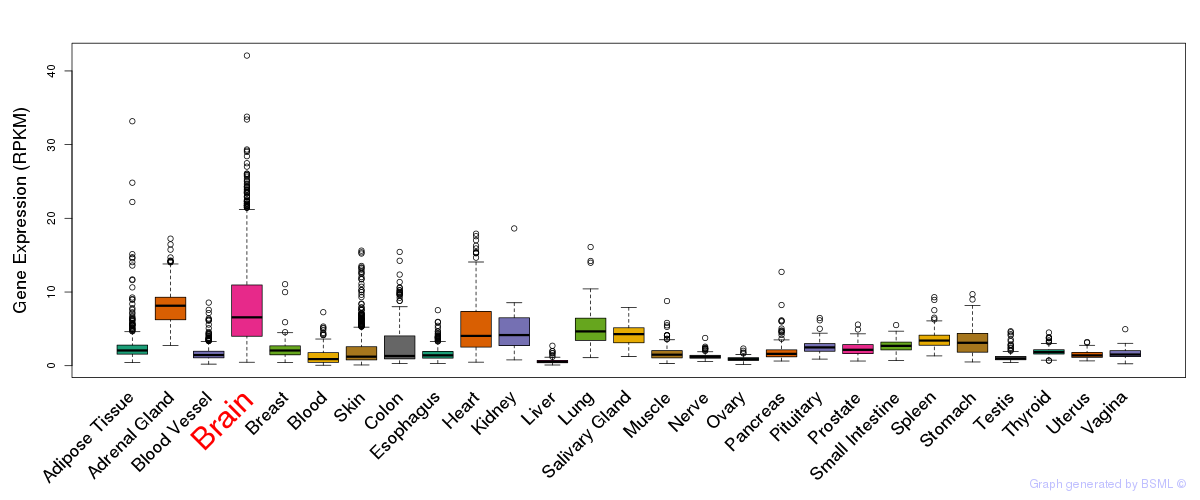
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PRKCZ | 0.88 | 0.89 |
| HMOX2 | 0.86 | 0.89 |
| PPP1R3F | 0.86 | 0.88 |
| ENTPD6 | 0.86 | 0.88 |
| ARHGEF4 | 0.85 | 0.84 |
| NRSN2 | 0.84 | 0.85 |
| SH3BP1 | 0.84 | 0.86 |
| MLF2 | 0.84 | 0.86 |
| CGREF1 | 0.84 | 0.84 |
| OTUB1 | 0.84 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.18 | -0.46 | -0.20 |
| AF347015.21 | -0.44 | -0.16 |
| AC010300.1 | -0.41 | -0.30 |
| MT-ATP8 | -0.39 | -0.15 |
| AF347015.2 | -0.38 | -0.13 |
| FAM159B | -0.38 | -0.46 |
| AP002478.3 | -0.38 | -0.28 |
| AF347015.8 | -0.38 | -0.16 |
| AF347015.26 | -0.37 | -0.13 |
| RAB13 | -0.36 | -0.41 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS RED UP | 17 | 13 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC2 TARGETS UP | 114 | 66 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| SANA TNF SIGNALING UP | 83 | 56 | All SZGR 2.0 genes in this pathway |
| KUNINGER IGF1 VS PDGFB TARGETS DN | 46 | 28 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE UP | 212 | 128 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION UP | 178 | 108 | All SZGR 2.0 genes in this pathway |
| IVANOVSKA MIR106B TARGETS | 90 | 56 | All SZGR 2.0 genes in this pathway |
| LINSLEY MIR16 TARGETS | 206 | 127 | All SZGR 2.0 genes in this pathway |
| BAKKER FOXO3 TARGETS UP | 61 | 41 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM SENSITIVE GENES | 90 | 54 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 3 TRANSIENTLY INDUCED BY EGF | 222 | 159 | All SZGR 2.0 genes in this pathway |