Gene Page: ALS2
Summary ?
| GeneID | 57679 |
| Symbol | ALS2 |
| Synonyms | ALS2CR6|ALSJ|IAHSP|PLSJ |
| Description | ALS2, alsin Rho guanine nucleotide exchange factor |
| Reference | MIM:606352|HGNC:HGNC:443|Ensembl:ENSG00000003393|HPRD:05893|Vega:OTTHUMG00000154507 |
| Gene type | protein-coding |
| Map location | 2q33.1 |
| Pascal p-value | 0.128 |
| Sherlock p-value | 0.107 |
| Fetal beta | 0.228 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 5 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01813165 | 2 | 202646406 | ALS2 | 8.85E-9 | -0.021 | 4.06E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | ALS2 | 57679 | 0.01 | trans | ||
| rs3845734 | chr2 | 171125572 | ALS2 | 57679 | 0.13 | trans | ||
| rs7584986 | chr2 | 184111432 | ALS2 | 57679 | 0.01 | trans | ||
| rs16955618 | chr15 | 29937543 | ALS2 | 57679 | 7.551E-7 | trans | ||
| rs1041786 | chr21 | 22617710 | ALS2 | 57679 | 0.02 | trans |
Section II. Transcriptome annotation
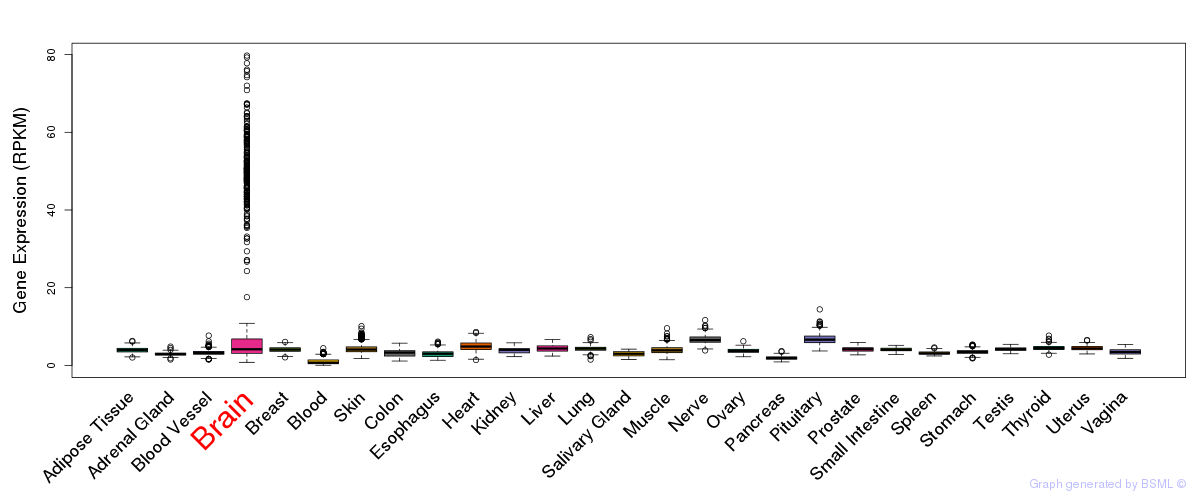
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005085 | guanyl-nucleotide exchange factor activity | IDA | 15247254 | |
| GO:0005087 | Ran guanyl-nucleotide exchange factor activity | NAS | 11586298 | |
| GO:0005515 | protein binding | IPI | 17239822 | |
| GO:0017112 | Rab guanyl-nucleotide exchange factor activity | IDA | 12837691 | |
| GO:0017137 | Rab GTPase binding | IDA | 15247254 | |
| GO:0017137 | Rab GTPase binding | NAS | 16670179 | |
| GO:0042803 | protein homodimerization activity | IPI | 15247254 | |
| GO:0030676 | Rac guanyl-nucleotide exchange factor activity | IDA | 16049005 | |
| GO:0043539 | protein serine/threonine kinase activator activity | IDA | 16049005 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007528 | neuromuscular junction development | IEA | Synap (GO term level: 10) | - |
| GO:0035249 | synaptic transmission, glutamatergic | IEA | neuron, glutamate, Synap, Neurotransmitter (GO term level: 8) | - |
| GO:0048812 | neurite morphogenesis | IDA | neuron, axon, neurite, dendrite (GO term level: 11) | 16049005 |
| GO:0001662 | behavioral fear response | IEA | - | |
| GO:0001881 | receptor recycling | IEA | - | |
| GO:0016050 | vesicle organization | IEA | - | |
| GO:0007626 | locomotory behavior | IEA | - | |
| GO:0008104 | protein localization | IEA | - | |
| GO:0006979 | response to oxidative stress | IEA | - | |
| GO:0007032 | endosome organization | NAS | 12837691 | |
| GO:0016197 | endosome transport | IEA | - | |
| GO:0035022 | positive regulation of Rac protein signal transduction | IC | 16049005 | |
| GO:0035023 | regulation of Rho protein signal transduction | IEA | - | |
| GO:0051036 | regulation of endosome size | IEP | 15247254 | |
| GO:0032313 | regulation of Rab GTPase activity | IEA | - | |
| GO:0032855 | positive regulation of Rac GTPase activity | IDA | 16049005 | |
| GO:0045860 | positive regulation of protein kinase activity | IDA | 16049005 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0014069 | postsynaptic density | IEA | Synap (GO term level: 10) | - |
| GO:0043197 | dendritic spine | IEA | Synap, dendrite (GO term level: 7) | - |
| GO:0001726 | ruffle | ISS | - | |
| GO:0005813 | centrosome | IDA | 16085057 | |
| GO:0005829 | cytosol | IDA | 12837691 | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005769 | early endosome | IDA | 12837691 | |
| GO:0030027 | lamellipodium | ISS | - | |
| GO:0042598 | vesicular fraction | IDA | 12837691 | |
| GO:0043234 | protein complex | IDA | 15247254 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG AMYOTROPHIC LATERAL SCLEROSIS ALS | 53 | 43 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP | 344 | 180 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| KEEN RESPONSE TO ROSIGLITAZONE UP | 38 | 23 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| COULOUARN TEMPORAL TGFB1 SIGNATURE UP | 109 | 68 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 TARGETS DN | 54 | 38 | All SZGR 2.0 genes in this pathway |
| GABRIELY MIR21 TARGETS | 289 | 187 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-142-5p | 1204 | 1211 | 1A,m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-144 | 1234 | 1240 | m8 | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-199 | 722 | 729 | 1A,m8 | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
| miR-26 | 448 | 455 | 1A,m8 | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-505 | 87 | 94 | 1A,m8 | hsa-miR-505 | GUCAACACUUGCUGGUUUCCUC |
| miR-543 | 1245 | 1251 | m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.