Gene Page: PTPRS
Summary ?
| GeneID | 5802 |
| Symbol | PTPRS |
| Synonyms | PTPSIGMA |
| Description | protein tyrosine phosphatase, receptor type S |
| Reference | MIM:601576|HGNC:HGNC:9681|Ensembl:ENSG00000105426|HPRD:03344|Vega:OTTHUMG00000180325 |
| Gene type | protein-coding |
| Map location | 19p13.3 |
| Pascal p-value | 0.174 |
| Fetal beta | 0.332 |
| DMG | 1 (# studies) |
| Support | TYROSINE KINASE SIGNALING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.2273 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09282805 | 19 | 5340122 | PTPRS | 6.96E-10 | -0.019 | 9.72E-7 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
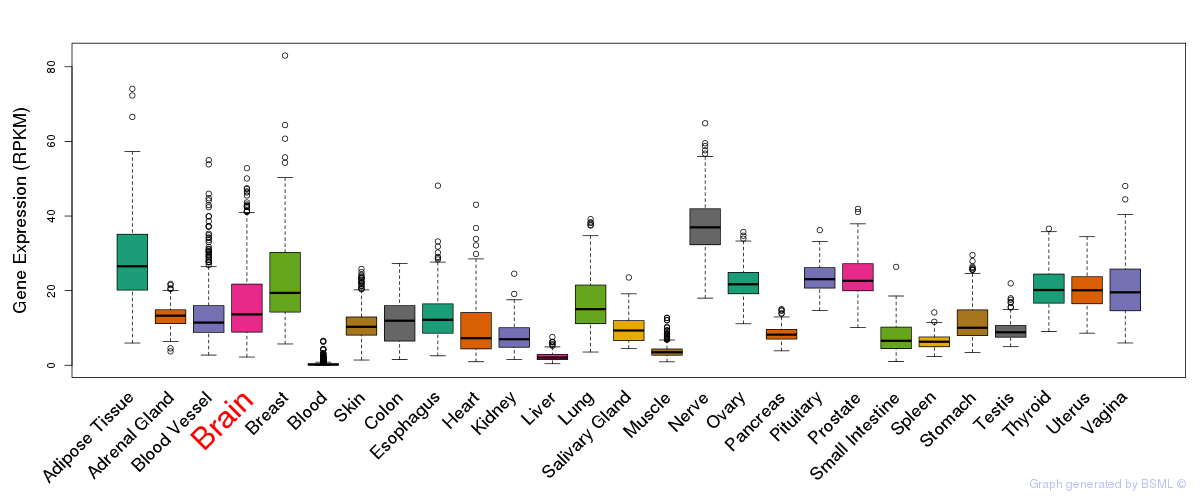
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TRAPPC9 | 0.86 | 0.84 |
| PSMD2 | 0.86 | 0.85 |
| VPS18 | 0.86 | 0.86 |
| DDB1 | 0.86 | 0.88 |
| DGCR2 | 0.86 | 0.85 |
| PFKL | 0.85 | 0.86 |
| P4HB | 0.84 | 0.84 |
| AP1M1 | 0.84 | 0.84 |
| VCP | 0.84 | 0.83 |
| ELAC2 | 0.84 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.73 | -0.77 |
| AF347015.21 | -0.73 | -0.77 |
| AF347015.31 | -0.73 | -0.75 |
| AF347015.33 | -0.70 | -0.73 |
| HIGD1B | -0.69 | -0.70 |
| MT-CYB | -0.69 | -0.72 |
| IFI27 | -0.68 | -0.68 |
| AF347015.8 | -0.68 | -0.73 |
| AF347015.27 | -0.68 | -0.73 |
| NOSTRIN | -0.66 | -0.67 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity | TAS | 8524829 | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 16273344 | |
| GO:0004725 | protein tyrosine phosphatase activity | IEA | - | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0016791 | phosphatase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007155 | cell adhesion | IEA | - | |
| GO:0006470 | protein amino acid dephosphorylation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 8524829 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CHD3 | Mi-2a | Mi2-ALPHA | ZFH | chromodomain helicase DNA binding protein 3 | Two-hybrid | BioGRID | 16169070 |
| EEF1G | EF1G | GIG35 | eukaryotic translation elongation factor 1 gamma | Two-hybrid | BioGRID | 16169070 |
| EIF4A2 | BM-010 | DDX2B | EIF4A | EIF4F | eukaryotic translation initiation factor 4A, isoform 2 | Affinity Capture-MS | BioGRID | 17353931 |
| FEZ1 | - | fasciculation and elongation protein zeta 1 (zygin I) | Two-hybrid | BioGRID | 16169070 |
| KAT5 | ESA1 | HTATIP | HTATIP1 | PLIP | TIP | TIP60 | cPLA2 | K(lysine) acetyltransferase 5 | Two-hybrid | BioGRID | 16169070 |
| KIAA1377 | - | KIAA1377 | Two-hybrid | BioGRID | 16169070 |
| MOBKL3 | 2C4D | CGI-95 | MGC12264 | MOB1 | MOB3 | PREI3 | MOB1, Mps One Binder kinase activator-like 3 (yeast) | Two-hybrid | BioGRID | 16169070 |
| PFN2 | D3S1319E | PFL | profilin 2 | Two-hybrid | BioGRID | 16169070 |
| PPFIA1 | FLJ41337 | FLJ42630 | FLJ43474 | LIP.1 | LIP1 | LIPRIN | MGC26800 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 | - | HPRD,BioGRID | 8524829 |
| PPFIA2 | FLJ41378 | MGC132572 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 | - | HPRD,BioGRID | 9624153 |
| PPFIA3 | KIAA0654 | LPNA3 | MGC126567 | MGC126569 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 | - | HPRD,BioGRID | 9624153 |
| PTN | HARP | HBGF8 | HBNF | NEGF1 | pleiotrophin | Two-hybrid | BioGRID | 16169070 |
| PTPRD | HPTP | HPTP-DELTA | HPTPD | MGC119750 | MGC119751 | MGC119752 | MGC119753 | PTPD | R-PTP-DELTA | protein tyrosine phosphatase, receptor type, D | - | HPRD,BioGRID | 9566880 |
| SETDB1 | ESET | KG1T | KIAA0067 | KMT1E | SET domain, bifurcated 1 | Two-hybrid | BioGRID | 16169070 |
| TUBB2A | TUBB | TUBB2 | dJ40E16.7 | tubulin, beta 2A | Two-hybrid | BioGRID | 16169070 |
| UBR1 | JBS | MGC142065 | MGC142067 | ubiquitin protein ligase E3 component n-recognin 1 | Two-hybrid | BioGRID | 16169070 |
| UNC119 | HRG4 | unc-119 homolog (C. elegans) | Two-hybrid | BioGRID | 16169070 |
| UTP14A | KIAA0266 | NY-CO-16 | SDCCAG16 | dJ537K23.3 | UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION DN | 100 | 64 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE UP | 134 | 93 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS MODERATELY UP | 109 | 69 | All SZGR 2.0 genes in this pathway |
| SUNG METASTASIS STROMA UP | 110 | 70 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| MOREAUX B LYMPHOCYTE MATURATION BY TACI UP | 92 | 58 | All SZGR 2.0 genes in this pathway |
| URS ADIPOCYTE DIFFERENTIATION UP | 74 | 51 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE UP | 149 | 85 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D UP | 139 | 95 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE DN | 72 | 47 | All SZGR 2.0 genes in this pathway |
| HOFFMANN SMALL PRE BII TO IMMATURE B LYMPHOCYTE DN | 50 | 33 | All SZGR 2.0 genes in this pathway |
| HOFMANN MYELODYSPLASTIC SYNDROM LOW RISK UP | 22 | 15 | All SZGR 2.0 genes in this pathway |
| HOFMANN MYELODYSPLASTIC SYNDROM HIGH RISK UP | 10 | 7 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT UP | 166 | 105 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 1 DN | 48 | 30 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 4 DN | 15 | 9 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-190 | 830 | 837 | 1A,m8 | hsa-miR-190 | UGAUAUGUUUGAUAUAUUAGGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.