Gene Page: PTPRZ1
Summary ?
| GeneID | 5803 |
| Symbol | PTPRZ1 |
| Synonyms | HPTPZ|HPTPzeta|PTP-ZETA|PTP18|PTPRZ|PTPZ|R-PTP-zeta-2|RPTPB|RPTPbeta|phosphacan |
| Description | protein tyrosine phosphatase, receptor type Z1 |
| Reference | MIM:176891|HGNC:HGNC:9685|Ensembl:ENSG00000106278|HPRD:01481|Vega:OTTHUMG00000157057 |
| Gene type | protein-coding |
| Map location | 7q31.3 |
| Pascal p-value | 0.261 |
| Sherlock p-value | 0.688 |
| Fetal beta | 1.666 |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 3 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
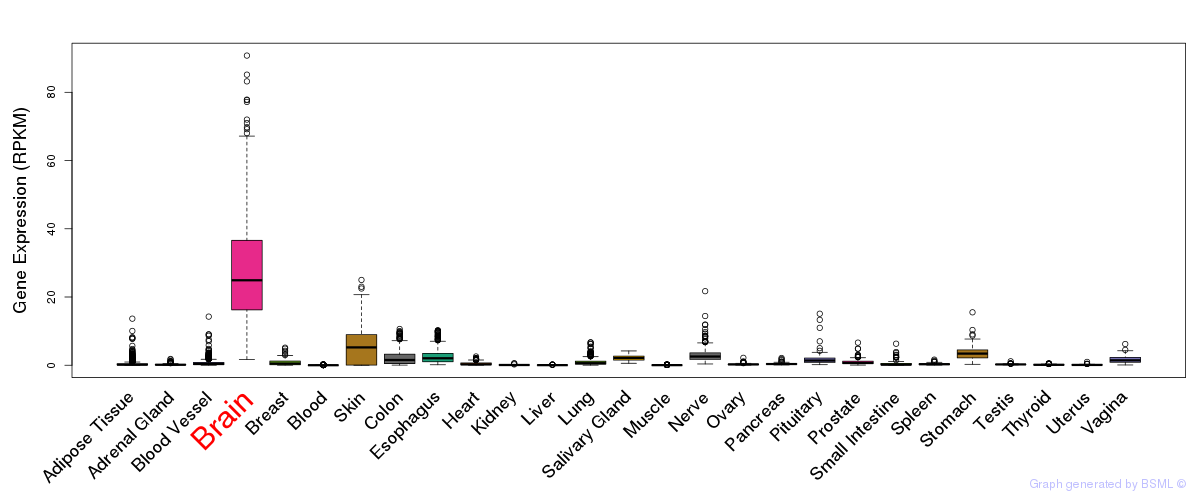
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity | TAS | 1323835 | |
| GO:0004089 | carbonate dehydratase activity | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008330 | protein tyrosine/threonine phosphatase activity | NAS | 9653645 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007417 | central nervous system development | TAS | Brain (GO term level: 6) | 8387522 |
| GO:0007409 | axonogenesis | IEA | neuron, axon, neurite (GO term level: 12) | - |
| GO:0006470 | protein amino acid dephosphorylation | TAS | 1323835 | |
| GO:0008150 | biological_process | ND | - | |
| GO:0006730 | one-carbon compound metabolic process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005575 | cellular_component | ND | - | |
| GO:0005578 | proteinaceous extracellular matrix | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 8387522 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG EPITHELIAL CELL SIGNALING IN HELICOBACTER PYLORI INFECTION | 68 | 44 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER DN | 169 | 118 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN | 258 | 141 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| KAN RESPONSE TO ARSENIC TRIOXIDE | 123 | 80 | All SZGR 2.0 genes in this pathway |
| GUENTHER GROWTH SPHERICAL VS ADHERENT UP | 21 | 15 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C8 | 72 | 56 | All SZGR 2.0 genes in this pathway |
| URS ADIPOCYTE DIFFERENTIATION UP | 74 | 51 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER DN | 101 | 65 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS MODEL DN | 43 | 28 | All SZGR 2.0 genes in this pathway |
| HUPER BREAST BASAL VS LUMINAL UP | 54 | 29 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS NEURAL UP | 18 | 13 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP | 198 | 128 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124/506 | 162 | 168 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-129-5p | 788 | 794 | m8 | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-133 | 512 | 518 | 1A | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| miR-200bc/429 | 449 | 455 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-221/222 | 354 | 360 | 1A | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
| miR-450 | 788 | 794 | 1A | hsa-miR-450 | UUUUUGCGAUGUGUUCCUAAUA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.