Gene Page: PEX5
Summary ?
| GeneID | 5830 |
| Symbol | PEX5 |
| Synonyms | PBD2A|PBD2B|PTS1-BP|PTS1R|PXR1|RCDP5 |
| Description | peroxisomal biogenesis factor 5 |
| Reference | MIM:600414|HGNC:HGNC:9719|Ensembl:ENSG00000139197|HPRD:02684|Vega:OTTHUMG00000168175 |
| Gene type | protein-coding |
| Map location | 12p13.31 |
| Pascal p-value | 0.05 |
| Sherlock p-value | 0.112 |
| Fetal beta | -0.319 |
| DMG | 3 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cortex Frontal Cortex BA9 Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg05236530 | 12 | 7362357 | PEX5 | 5.888E-4 | 0.364 | 0.05 | DMG:Wockner_2014 |
| cg15754084 | 12 | 7343020 | PEX5 | -0.038 | 0.87 | DMG:Nishioka_2013 | |
| cg07748017 | 12 | 7342621 | PEX5 | 5.83E-8 | -0.012 | 1.49E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2456470 | chr15 | 62777955 | PEX5 | 5830 | 0.19 | trans | ||
| rs2053887 | 12 | 7346530 | PEX5 | ENSG00000139197.6 | 1.478E-6 | 0.01 | 5249 | gtex_brain_ba24 |
| rs12830671 | 12 | 7347278 | PEX5 | ENSG00000139197.6 | 8.763E-7 | 0.01 | 5997 | gtex_brain_ba24 |
| rs12820829 | 12 | 7347534 | PEX5 | ENSG00000139197.6 | 8.763E-7 | 0.01 | 6253 | gtex_brain_ba24 |
| rs61917956 | 12 | 7348659 | PEX5 | ENSG00000139197.6 | 4.544E-7 | 0.01 | 7378 | gtex_brain_ba24 |
| rs78613188 | 12 | 7348728 | PEX5 | ENSG00000139197.6 | 9.674E-8 | 0.01 | 7447 | gtex_brain_ba24 |
| rs200498082 | 12 | 7348907 | PEX5 | ENSG00000139197.6 | 1.518E-7 | 0.01 | 7626 | gtex_brain_ba24 |
| rs61917957 | 12 | 7349987 | PEX5 | ENSG00000139197.6 | 7.538E-7 | 0.01 | 8706 | gtex_brain_ba24 |
| rs200609395 | 12 | 7350135 | PEX5 | ENSG00000139197.6 | 7.415E-8 | 0.01 | 8854 | gtex_brain_ba24 |
| rs185073397 | 12 | 7350136 | PEX5 | ENSG00000139197.6 | 5.77E-7 | 0.01 | 8855 | gtex_brain_ba24 |
| rs7139158 | 12 | 7350155 | PEX5 | ENSG00000139197.6 | 1.011E-7 | 0.01 | 8874 | gtex_brain_ba24 |
| rs7962563 | 12 | 7353514 | PEX5 | ENSG00000139197.6 | 8.647E-7 | 0.01 | 12233 | gtex_brain_ba24 |
| rs10743271 | 12 | 7353790 | PEX5 | ENSG00000139197.6 | 1.326E-7 | 0.01 | 12509 | gtex_brain_ba24 |
| rs7315553 | 12 | 7353838 | PEX5 | ENSG00000139197.6 | 8.667E-7 | 0.01 | 12557 | gtex_brain_ba24 |
| rs12825629 | 12 | 7355583 | PEX5 | ENSG00000139197.6 | 8.702E-7 | 0.01 | 14302 | gtex_brain_ba24 |
| rs6486910 | 12 | 7356479 | PEX5 | ENSG00000139197.6 | 3.866E-7 | 0.01 | 15198 | gtex_brain_ba24 |
| rs6486911 | 12 | 7356494 | PEX5 | ENSG00000139197.6 | 3.874E-7 | 0.01 | 15213 | gtex_brain_ba24 |
| rs7970916 | 12 | 7357746 | PEX5 | ENSG00000139197.6 | 8.763E-7 | 0.01 | 16465 | gtex_brain_ba24 |
| rs75912799 | 12 | 7357897 | PEX5 | ENSG00000139197.6 | 8.763E-7 | 0.01 | 16616 | gtex_brain_ba24 |
| rs10161405 | 12 | 7358646 | PEX5 | ENSG00000139197.6 | 3.071E-7 | 0.01 | 17365 | gtex_brain_ba24 |
| rs10161542 | 12 | 7358684 | PEX5 | ENSG00000139197.6 | 3.071E-7 | 0.01 | 17403 | gtex_brain_ba24 |
| rs11044332 | 12 | 7358768 | PEX5 | ENSG00000139197.6 | 8.763E-7 | 0.01 | 17487 | gtex_brain_ba24 |
| rs111408486 | 12 | 7359175 | PEX5 | ENSG00000139197.6 | 8.763E-7 | 0.01 | 17894 | gtex_brain_ba24 |
| rs397737333 | 12 | 7359309 | PEX5 | ENSG00000139197.6 | 8.763E-7 | 0.01 | 18028 | gtex_brain_ba24 |
| rs3825405 | 12 | 7360143 | PEX5 | ENSG00000139197.6 | 8.763E-7 | 0.01 | 18862 | gtex_brain_ba24 |
| rs3816424 | 12 | 7360229 | PEX5 | ENSG00000139197.6 | 1.42E-7 | 0.01 | 18948 | gtex_brain_ba24 |
| rs1057225 | 12 | 7363574 | PEX5 | ENSG00000139197.6 | 8.763E-7 | 0.01 | 22293 | gtex_brain_ba24 |
| rs11448434 | 12 | 7363943 | PEX5 | ENSG00000139197.6 | 1.015E-8 | 0.01 | 22662 | gtex_brain_ba24 |
| rs4329748 | 12 | 7364442 | PEX5 | ENSG00000139197.6 | 8.763E-7 | 0.01 | 23161 | gtex_brain_ba24 |
| rs4492891 | 12 | 7364789 | PEX5 | ENSG00000139197.6 | 1.497E-6 | 0.01 | 23508 | gtex_brain_ba24 |
| rs200081407 | 12 | 7366581 | PEX5 | ENSG00000139197.6 | 8.857E-7 | 0.01 | 25300 | gtex_brain_ba24 |
| rs12228393 | 12 | 7368182 | PEX5 | ENSG00000139197.6 | 8.763E-7 | 0.01 | 26901 | gtex_brain_ba24 |
| rs10161170 | 12 | 7368629 | PEX5 | ENSG00000139197.6 | 1.422E-7 | 0.01 | 27348 | gtex_brain_ba24 |
| rs11044901 | 12 | 7368691 | PEX5 | ENSG00000139197.6 | 1.413E-7 | 0.01 | 27410 | gtex_brain_ba24 |
| rs10161103 | 12 | 7369087 | PEX5 | ENSG00000139197.6 | 1.42E-7 | 0.01 | 27806 | gtex_brain_ba24 |
| rs7969508 | 12 | 7370502 | PEX5 | ENSG00000139197.6 | 1.427E-7 | 0.01 | 29221 | gtex_brain_ba24 |
| rs7969635 | 12 | 7370612 | PEX5 | ENSG00000139197.6 | 1.427E-7 | 0.01 | 29331 | gtex_brain_ba24 |
| rs7969751 | 12 | 7370702 | PEX5 | ENSG00000139197.6 | 1.423E-7 | 0.01 | 29421 | gtex_brain_ba24 |
| rs7962917 | 12 | 7317194 | PEX5 | ENSG00000139197.6 | 1.276E-8 | 0 | -24087 | gtex_brain_putamen_basal |
| rs1450962 | 12 | 7320545 | PEX5 | ENSG00000139197.6 | 2.133E-10 | 0 | -20736 | gtex_brain_putamen_basal |
| rs12828040 | 12 | 7326541 | PEX5 | ENSG00000139197.6 | 1.997E-10 | 0 | -14740 | gtex_brain_putamen_basal |
| rs7310539 | 12 | 7329996 | PEX5 | ENSG00000139197.6 | 2.435E-10 | 0 | -11285 | gtex_brain_putamen_basal |
| rs111377129 | 12 | 7331606 | PEX5 | ENSG00000139197.6 | 1.413E-6 | 0 | -9675 | gtex_brain_putamen_basal |
| rs1839622 | 12 | 7334534 | PEX5 | ENSG00000139197.6 | 2.247E-10 | 0 | -6747 | gtex_brain_putamen_basal |
| rs7973719 | 12 | 7334813 | PEX5 | ENSG00000139197.6 | 6.325E-10 | 0 | -6468 | gtex_brain_putamen_basal |
| rs12828421 | 12 | 7335217 | PEX5 | ENSG00000139197.6 | 2.079E-10 | 0 | -6064 | gtex_brain_putamen_basal |
| rs12816419 | 12 | 7335420 | PEX5 | ENSG00000139197.6 | 6.331E-10 | 0 | -5861 | gtex_brain_putamen_basal |
| rs397742110 | 12 | 7336926 | PEX5 | ENSG00000139197.6 | 5.252E-10 | 0 | -4355 | gtex_brain_putamen_basal |
| rs397851127 | 12 | 7338793 | PEX5 | ENSG00000139197.6 | 9.714E-9 | 0 | -2488 | gtex_brain_putamen_basal |
| rs61917955 | 12 | 7338947 | PEX5 | ENSG00000139197.6 | 1.839E-10 | 0 | -2334 | gtex_brain_putamen_basal |
| rs12227917 | 12 | 7342343 | PEX5 | ENSG00000139197.6 | 2.655E-10 | 0 | 1062 | gtex_brain_putamen_basal |
| rs200807978 | 12 | 7342364 | PEX5 | ENSG00000139197.6 | 3.05E-9 | 0 | 1083 | gtex_brain_putamen_basal |
| rs201786247 | 12 | 7342367 | PEX5 | ENSG00000139197.6 | 1.249E-6 | 0 | 1086 | gtex_brain_putamen_basal |
| rs200032511 | 12 | 7342371 | PEX5 | ENSG00000139197.6 | 6.54E-7 | 0 | 1090 | gtex_brain_putamen_basal |
| rs12424618 | 12 | 7344802 | PEX5 | ENSG00000139197.6 | 1.258E-11 | 0 | 3521 | gtex_brain_putamen_basal |
| rs2053887 | 12 | 7346530 | PEX5 | ENSG00000139197.6 | 4.144E-12 | 0 | 5249 | gtex_brain_putamen_basal |
| rs12830671 | 12 | 7347278 | PEX5 | ENSG00000139197.6 | 2.807E-12 | 0 | 5997 | gtex_brain_putamen_basal |
| rs12820829 | 12 | 7347534 | PEX5 | ENSG00000139197.6 | 2.799E-12 | 0 | 6253 | gtex_brain_putamen_basal |
| rs61917956 | 12 | 7348659 | PEX5 | ENSG00000139197.6 | 3.767E-12 | 0 | 7378 | gtex_brain_putamen_basal |
| rs78613188 | 12 | 7348728 | PEX5 | ENSG00000139197.6 | 1.218E-11 | 0 | 7447 | gtex_brain_putamen_basal |
| rs200897876 | 12 | 7348841 | PEX5 | ENSG00000139197.6 | 1.086E-8 | 0 | 7560 | gtex_brain_putamen_basal |
| rs200264446 | 12 | 7348912 | PEX5 | ENSG00000139197.6 | 2.823E-7 | 0 | 7631 | gtex_brain_putamen_basal |
| rs181450002 | 12 | 7349108 | PEX5 | ENSG00000139197.6 | 1.562E-8 | 0 | 7827 | gtex_brain_putamen_basal |
| rs186088555 | 12 | 7349135 | PEX5 | ENSG00000139197.6 | 3.921E-7 | 0 | 7854 | gtex_brain_putamen_basal |
| rs183222544 | 12 | 7349162 | PEX5 | ENSG00000139197.6 | 1.979E-9 | 0 | 7881 | gtex_brain_putamen_basal |
| rs61917957 | 12 | 7349987 | PEX5 | ENSG00000139197.6 | 2.169E-9 | 0 | 8706 | gtex_brain_putamen_basal |
| rs71067161 | 12 | 7350013 | PEX5 | ENSG00000139197.6 | 7.964E-10 | 0 | 8732 | gtex_brain_putamen_basal |
| rs117028872 | 12 | 7350028 | PEX5 | ENSG00000139197.6 | 1.328E-6 | 0 | 8747 | gtex_brain_putamen_basal |
| rs79081846 | 12 | 7350052 | PEX5 | ENSG00000139197.6 | 9.669E-10 | 0 | 8771 | gtex_brain_putamen_basal |
| rs200609395 | 12 | 7350135 | PEX5 | ENSG00000139197.6 | 3.462E-8 | 0 | 8854 | gtex_brain_putamen_basal |
| rs185073397 | 12 | 7350136 | PEX5 | ENSG00000139197.6 | 5.01E-7 | 0 | 8855 | gtex_brain_putamen_basal |
| rs7139158 | 12 | 7350155 | PEX5 | ENSG00000139197.6 | 2.458E-8 | 0 | 8874 | gtex_brain_putamen_basal |
| rs78964892 | 12 | 7352735 | PEX5 | ENSG00000139197.6 | 6.008E-10 | 0 | 11454 | gtex_brain_putamen_basal |
| rs138686752 | 12 | 7352903 | PEX5 | ENSG00000139197.6 | 2.405E-12 | 0 | 11622 | gtex_brain_putamen_basal |
| rs7973600 | 12 | 7353351 | PEX5 | ENSG00000139197.6 | 6.853E-14 | 0 | 12070 | gtex_brain_putamen_basal |
| rs7962563 | 12 | 7353514 | PEX5 | ENSG00000139197.6 | 1.329E-13 | 0 | 12233 | gtex_brain_putamen_basal |
| rs10743271 | 12 | 7353790 | PEX5 | ENSG00000139197.6 | 2.402E-10 | 0 | 12509 | gtex_brain_putamen_basal |
| rs7315553 | 12 | 7353838 | PEX5 | ENSG00000139197.6 | 1.455E-13 | 0 | 12557 | gtex_brain_putamen_basal |
| rs12825629 | 12 | 7355583 | PEX5 | ENSG00000139197.6 | 1.005E-13 | 0 | 14302 | gtex_brain_putamen_basal |
| rs6486910 | 12 | 7356479 | PEX5 | ENSG00000139197.6 | 1.906E-13 | 0 | 15198 | gtex_brain_putamen_basal |
| rs6486911 | 12 | 7356494 | PEX5 | ENSG00000139197.6 | 1.904E-13 | 0 | 15213 | gtex_brain_putamen_basal |
| rs372666240 | 12 | 7356574 | PEX5 | ENSG00000139197.6 | 1.163E-6 | 0 | 15293 | gtex_brain_putamen_basal |
| rs7970916 | 12 | 7357746 | PEX5 | ENSG00000139197.6 | 1.449E-13 | 0 | 16465 | gtex_brain_putamen_basal |
| rs75912799 | 12 | 7357897 | PEX5 | ENSG00000139197.6 | 1.449E-13 | 0 | 16616 | gtex_brain_putamen_basal |
| rs10161405 | 12 | 7358646 | PEX5 | ENSG00000139197.6 | 5.601E-8 | 0 | 17365 | gtex_brain_putamen_basal |
| rs10161542 | 12 | 7358684 | PEX5 | ENSG00000139197.6 | 5.601E-8 | 0 | 17403 | gtex_brain_putamen_basal |
| rs11044332 | 12 | 7358768 | PEX5 | ENSG00000139197.6 | 1.449E-13 | 0 | 17487 | gtex_brain_putamen_basal |
| rs111408486 | 12 | 7359175 | PEX5 | ENSG00000139197.6 | 1.449E-13 | 0 | 17894 | gtex_brain_putamen_basal |
| rs397737333 | 12 | 7359309 | PEX5 | ENSG00000139197.6 | 1.449E-13 | 0 | 18028 | gtex_brain_putamen_basal |
| rs3825405 | 12 | 7360143 | PEX5 | ENSG00000139197.6 | 1.449E-13 | 0 | 18862 | gtex_brain_putamen_basal |
| rs3816424 | 12 | 7360229 | PEX5 | ENSG00000139197.6 | 6.524E-10 | 0 | 18948 | gtex_brain_putamen_basal |
| rs1057225 | 12 | 7363574 | PEX5 | ENSG00000139197.6 | 1.449E-13 | 0 | 22293 | gtex_brain_putamen_basal |
| rs11448434 | 12 | 7363943 | PEX5 | ENSG00000139197.6 | 1.995E-9 | 0 | 22662 | gtex_brain_putamen_basal |
| rs4329748 | 12 | 7364442 | PEX5 | ENSG00000139197.6 | 1.449E-13 | 0 | 23161 | gtex_brain_putamen_basal |
| rs4512930 | 12 | 7364623 | PEX5 | ENSG00000139197.6 | 2.205E-13 | 0 | 23342 | gtex_brain_putamen_basal |
| rs4492891 | 12 | 7364789 | PEX5 | ENSG00000139197.6 | 1.454E-13 | 0 | 23508 | gtex_brain_putamen_basal |
| rs200081407 | 12 | 7366581 | PEX5 | ENSG00000139197.6 | 1.143E-9 | 0 | 25300 | gtex_brain_putamen_basal |
| rs12228393 | 12 | 7368182 | PEX5 | ENSG00000139197.6 | 2.807E-12 | 0 | 26901 | gtex_brain_putamen_basal |
| rs10161170 | 12 | 7368629 | PEX5 | ENSG00000139197.6 | 6.543E-10 | 0 | 27348 | gtex_brain_putamen_basal |
| rs11044901 | 12 | 7368691 | PEX5 | ENSG00000139197.6 | 6.612E-10 | 0 | 27410 | gtex_brain_putamen_basal |
| rs10161103 | 12 | 7369087 | PEX5 | ENSG00000139197.6 | 6.524E-10 | 0 | 27806 | gtex_brain_putamen_basal |
| rs397830029 | 12 | 7369169 | PEX5 | ENSG00000139197.6 | 1.22E-12 | 0 | 27888 | gtex_brain_putamen_basal |
| rs6487053 | 12 | 7369318 | PEX5 | ENSG00000139197.6 | 1.275E-12 | 0 | 28037 | gtex_brain_putamen_basal |
| rs61919598 | 12 | 7370116 | PEX5 | ENSG00000139197.6 | 5.956E-7 | 0 | 28835 | gtex_brain_putamen_basal |
| rs7969508 | 12 | 7370502 | PEX5 | ENSG00000139197.6 | 7.99E-10 | 0 | 29221 | gtex_brain_putamen_basal |
| rs7969635 | 12 | 7370612 | PEX5 | ENSG00000139197.6 | 7.99E-10 | 0 | 29331 | gtex_brain_putamen_basal |
| rs7969751 | 12 | 7370702 | PEX5 | ENSG00000139197.6 | 8E-10 | 0 | 29421 | gtex_brain_putamen_basal |
| rs72098838 | 12 | 7371483 | PEX5 | ENSG00000139197.6 | 2.216E-7 | 0 | 30202 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
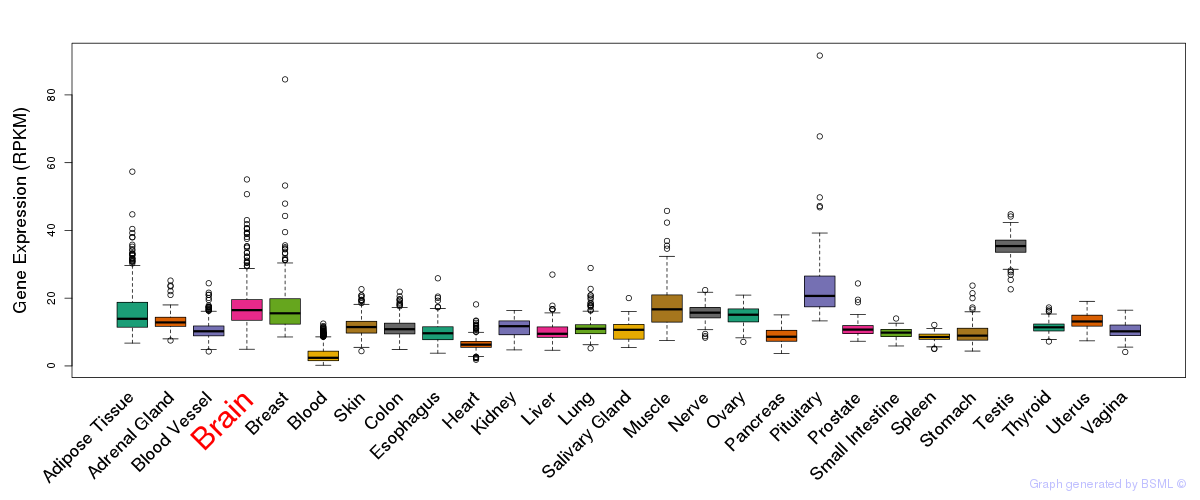
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ATAD1 | 0.93 | 0.91 |
| SLC30A9 | 0.92 | 0.91 |
| RAB18 | 0.92 | 0.92 |
| CUL2 | 0.92 | 0.89 |
| STX12 | 0.92 | 0.91 |
| FAM49B | 0.92 | 0.90 |
| USP33 | 0.92 | 0.90 |
| C10orf88 | 0.91 | 0.90 |
| PPP2CA | 0.91 | 0.89 |
| CCDC132 | 0.91 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.74 | -0.68 |
| AF347015.33 | -0.72 | -0.66 |
| AF347015.8 | -0.71 | -0.67 |
| MT-CYB | -0.71 | -0.66 |
| FXYD1 | -0.71 | -0.68 |
| AF347015.2 | -0.71 | -0.65 |
| AF347015.26 | -0.71 | -0.66 |
| HIGD1B | -0.69 | -0.64 |
| AF347015.31 | -0.69 | -0.65 |
| AF347015.21 | -0.69 | -0.60 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 9653144 |10837480 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001764 | neuron migration | IEA | neuron (GO term level: 8) | - |
| GO:0021895 | cerebral cortex neuron differentiation | IEA | neuron, Brain (GO term level: 9) | - |
| GO:0000038 | very-long-chain fatty acid metabolic process | IEA | - | |
| GO:0007006 | mitochondrial membrane organization | IEA | - | |
| GO:0048468 | cell development | IEA | - | |
| GO:0007031 | peroxisome organization | IEA | - | |
| GO:0007029 | endoplasmic reticulum organization | IEA | - | |
| GO:0006635 | fatty acid beta-oxidation | IEA | - | |
| GO:0015031 | protein transport | IEA | - | |
| GO:0040018 | positive regulation of multicellular organism growth | IEA | - | |
| GO:0021795 | cerebral cortex cell migration | IEA | - | |
| GO:0044255 | cellular lipid metabolic process | IEA | - | |
| GO:0050905 | neuromuscular process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005777 | peroxisome | IEA | - | |
| GO:0005778 | peroxisomal membrane | IEA | - | |
| GO:0016020 | membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATM | AT1 | ATA | ATC | ATD | ATDC | ATE | DKFZp781A0353 | MGC74674 | TEL1 | TELO1 | ataxia telangiectasia mutated | - | HPRD,BioGRID | 10567403 |
| CAT | MGC138422 | MGC138424 | catalase | Two-hybrid | BioGRID | 10567403 |
| DDO | DASOX | DDO-1 | DDO-2 | FLJ45203 | D-aspartate oxidase | - | HPRD,BioGRID | 9820813 |
| HSPA1A | FLJ54303 | FLJ54370 | FLJ54392 | FLJ54408 | FLJ75127 | HSP70-1 | HSP70-1A | HSP70I | HSP72 | HSPA1 | HSPA1B | heat shock 70kDa protein 1A | - | HPRD | 11415446 |
| PDZK1 | CAP70 | CLAMP | NHERF3 | PDZD1 | PDZ domain containing 1 | Two-hybrid | BioGRID | 16189514 |
| PEX10 | MGC1998 | NALD | RNF69 | peroxisomal biogenesis factor 10 | - | HPRD,BioGRID | 10837480 |
| PEX12 | PAF-3 | peroxisomal biogenesis factor 12 | PEX12 interacts with PEX5. | BIND | 12096124 |
| PEX12 | PAF-3 | peroxisomal biogenesis factor 12 | - | HPRD,BioGRID | 10562279 |10837480 |
| PEX13 | NALD | ZWS | peroxisomal biogenesis factor 13 | - | HPRD,BioGRID | 8858165 |11865044 |
| PEX14 | MGC12767 | NAPP2 | Pex14p | dJ734G22.2 | peroxisomal biogenesis factor 14 | PEX14 interacts with PEX5. | BIND | 12096124 |
| PEX14 | MGC12767 | NAPP2 | Pex14p | dJ734G22.2 | peroxisomal biogenesis factor 14 | Reconstituted Complex Two-hybrid | BioGRID | 10212238 |10837480 |11865044 |
| PEX14 | MGC12767 | NAPP2 | Pex14p | dJ734G22.2 | peroxisomal biogenesis factor 14 | - | HPRD | 10212238 |11865044 |
| PEX7 | PTS2R | RCDP1 | RD | peroxisomal biogenesis factor 7 | - | HPRD,BioGRID | 11546814 |11865044 |
| PXMP3 | PAF-1 | PAF1 | PEX2 | PMP3 | PMP35 | RNF72 | peroxisomal membrane protein 3, 35kDa | - | HPRD | 10837480 |
| SIRT3 | SIR2L3 | sirtuin (silent mating type information regulation 2 homolog) 3 (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| SNUPN | KPNBL | RNUT1 | Snurportin1 | snurportin 1 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PEROXISOME | 78 | 47 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN DN | 241 | 146 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED DN | 86 | 47 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL UP | 133 | 78 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 5 | 147 | 89 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| ZHAN V2 LATE DIFFERENTIATION GENES | 45 | 34 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION MUSCLE UP | 43 | 33 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| YOSHIOKA LIVER CANCER EARLY RECURRENCE UP | 40 | 23 | All SZGR 2.0 genes in this pathway |
| ACOSTA PROLIFERATION INDEPENDENT MYC TARGETS UP | 84 | 51 | All SZGR 2.0 genes in this pathway |
| TERAO AOX4 TARGETS HG UP | 28 | 20 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND WITH H4K20ME1 MARK | 145 | 82 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-141/200a | 919 | 926 | 1A,m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-15/16/195/424/497 | 272 | 278 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-29 | 269 | 275 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-31 | 892 | 899 | 1A,m8 | hsa-miR-31 | AGGCAAGAUGCUGGCAUAGCUG |
| miR-361 | 330 | 336 | m8 | hsa-miR-361brain | UUAUCAGAAUCUCCAGGGGUAC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.