Gene Page: PZP
Summary ?
| GeneID | 5858 |
| Symbol | PZP |
| Synonyms | CPAMD6 |
| Description | pregnancy-zone protein |
| Reference | MIM:176420|HGNC:HGNC:9750|Ensembl:ENSG00000126838|HPRD:07178|Vega:OTTHUMG00000154915 |
| Gene type | protein-coding |
| Map location | 12p13-p12.2 |
| Pascal p-value | 0.534 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
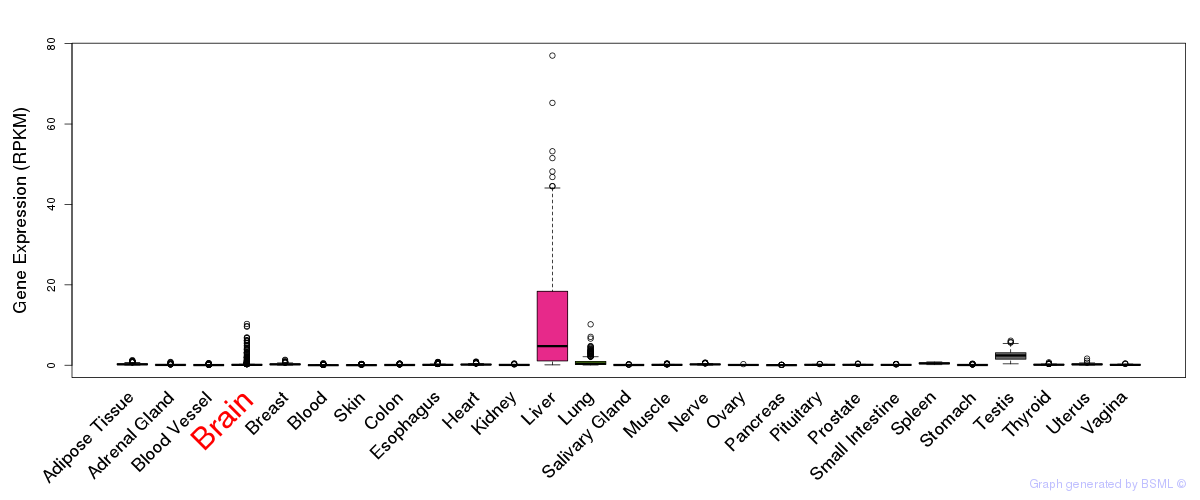
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SEPT2 | 0.71 | 0.61 |
| CRB1 | 0.71 | 0.53 |
| HEATR5A | 0.70 | 0.55 |
| SEPT10 | 0.70 | 0.56 |
| NEDD1 | 0.69 | 0.45 |
| HAT1 | 0.69 | 0.44 |
| YAP1 | 0.69 | 0.53 |
| NUP37 | 0.68 | 0.46 |
| KIF24 | 0.67 | 0.44 |
| C5 | 0.67 | 0.46 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RTN4RL1 | -0.25 | -0.20 |
| ANKRD24 | -0.25 | -0.18 |
| SLC26A4 | -0.24 | -0.22 |
| CACNA1A | -0.24 | -0.19 |
| ARPP-21 | -0.23 | -0.20 |
| LRFN2 | -0.23 | -0.20 |
| FAM19A1 | -0.22 | -0.18 |
| HIVEP3 | -0.22 | -0.19 |
| ACTN2 | -0.22 | -0.15 |
| AC217771.1 | -0.21 | -0.18 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 24HR DN | 33 | 21 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND ENDOTHELIUM | 66 | 47 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA DN | 267 | 160 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| NABA ECM REGULATORS | 238 | 125 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |