Gene Page: TRIM27
Summary ?
| GeneID | 5987 |
| Symbol | TRIM27 |
| Synonyms | RFP|RNF76 |
| Description | tripartite motif containing 27 |
| Reference | MIM:602165|HGNC:HGNC:9975|Ensembl:ENSG00000204713|HPRD:15996|Vega:OTTHUMG00000031215 |
| Gene type | protein-coding |
| Map location | 6p22 |
| Pascal p-value | 1E-12 |
| Sherlock p-value | 0.72 |
| Fetal beta | 0.756 |
| DMG | 2 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg11629443 | 6 | 28891340 | TRIM27 | 1.53E-5 | -0.512 | 0.015 | DMG:Wockner_2014 |
| cg26135345 | 6 | 28806907 | TRIM27 | 4.18E-9 | -0.016 | 2.51E-6 | DMG:Jaffe_2016 |
| cg10162774 | 6 | 28864168 | TRIM27 | 3.33E-8 | -0.019 | 9.98E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| snp_a-1856993 | 0 | TRIM27 | 5987 | 0.18 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
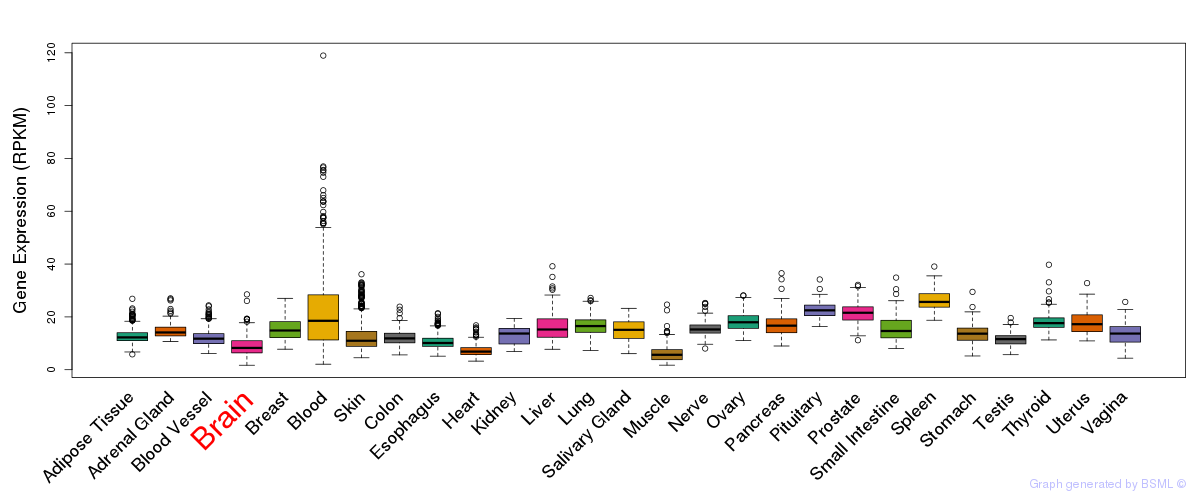
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PDLIM3 | 0.52 | 0.43 |
| PECI | 0.50 | 0.44 |
| CD99 | 0.50 | 0.35 |
| DBI | 0.49 | 0.43 |
| NUP37 | 0.47 | 0.35 |
| SNRPG | 0.46 | 0.31 |
| TEX9 | 0.46 | 0.21 |
| EFNA4 | 0.45 | 0.35 |
| CLIC1 | 0.45 | 0.34 |
| RAB32 | 0.45 | 0.30 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NDRG1 | -0.35 | -0.40 |
| SLCO3A1 | -0.35 | -0.37 |
| AC016910.1 | -0.35 | -0.39 |
| DOCK9 | -0.35 | -0.34 |
| C1orf115 | -0.34 | -0.39 |
| KIF16B | -0.34 | -0.30 |
| SPTBN2 | -0.33 | -0.34 |
| RHOU | -0.33 | -0.37 |
| SPTBN1 | -0.33 | -0.32 |
| DUSP7 | -0.33 | -0.35 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004714 | transmembrane receptor protein tyrosine kinase activity | TAS | neurite (GO term level: 8) | 3078962 |
| GO:0003677 | DNA binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 9570750 |15837424 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | TAS | 3380101 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | IDA | 10976108 | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0007283 | spermatogenesis | TAS | 3380101 | |
| GO:0008283 | cell proliferation | TAS | 2734021 | |
| GO:0045814 | negative regulation of gene expression, epigenetic | IDA | 10976108 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005624 | membrane fraction | TAS | 3078962 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | IDA | 10976108 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 3078962 | |
| GO:0031965 | nuclear membrane | IDA | 10976108 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BEGAIN | KIAA1446 | brain-enriched guanylate kinase-associated homolog (rat) | Two-hybrid | BioGRID | 16189514 |
| CHD4 | DKFZp686E06161 | Mi-2b | Mi2-BETA | chromodomain helicase DNA binding protein 4 | - | HPRD | 14530259 |
| DNMT1 | AIM | CXXC9 | DNMT | FLJ16293 | MCMT | MGC104992 | DNA (cytosine-5-)-methyltransferase 1 | Two-hybrid | BioGRID | 16169070 |
| EIF3E | EIF3-P48 | EIF3S6 | INT6 | eIF3-p46 | eukaryotic translation initiation factor 3, subunit E | - | HPRD | 10504338 |
| EPC1 | DKFZp781P2312 | Epl1 | enhancer of polycomb homolog 1 (Drosophila) | - | HPRD | 10976108 |
| FBF1 | Alb | FBF-1 | FLJ00103 | Fas (TNFRSF6) binding factor 1 | Two-hybrid | BioGRID | 16189514 |
| FXYD6 | - | FXYD domain containing ion transport regulator 6 | Two-hybrid | BioGRID | 16169070 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD | 14530259 |
| MAGEA11 | MAGE-11 | MAGE11 | MAGEA-11 | MGC10511 | melanoma antigen family A, 11 | Two-hybrid | BioGRID | 16189514 |
| MCOLN1 | ML4 | MLIV | MST080 | MSTP080 | TRP-ML1 | TRPM-L1 | TRPML1 | mucolipin 1 | Two-hybrid | BioGRID | 16169070 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | - | HPRD,BioGRID | 9570750 |
| PRAM1 | MGC39864 | PRAM-1 | PML-RARA regulated adaptor molecule 1 | Affinity Capture-Western Two-hybrid | BioGRID | 9570750 |
| TRIM11 | BIA1 | RNF92 | tripartite motif-containing 11 | - | HPRD,BioGRID | 11331580 |
| TRIM23 | ARD1 | ARFD1 | RNF46 | tripartite motif-containing 23 | - | HPRD,BioGRID | 11331580 |
| TRIM27 | RFP | RNF76 | tripartite motif-containing 27 | Two-hybrid | BioGRID | 11331580 |
| TRIM29 | ATDC | FLJ36085 | tripartite motif-containing 29 | - | HPRD,BioGRID | 11331580 |
| TRIM32 | BBS11 | HT2A | LGMD2H | TATIP | tripartite motif-containing 32 | - | HPRD,BioGRID | 11331580 |
| ZBTB16 | PLZF | ZNF145 | zinc finger and BTB domain containing 16 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| CASORELLI APL SECONDARY VS DE NOVO UP | 39 | 25 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION | 77 | 51 | All SZGR 2.0 genes in this pathway |
| SCHEIDEREIT IKK INTERACTING PROTEINS | 58 | 45 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM UP | 176 | 111 | All SZGR 2.0 genes in this pathway |
| TOMLINS PROSTATE CANCER UP | 40 | 27 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| HOWLIN CITED1 TARGETS 2 UP | 17 | 10 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 2G | 171 | 96 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BAKKER FOXO3 TARGETS DN | 187 | 109 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL DN | 146 | 88 | All SZGR 2.0 genes in this pathway |