Gene Page: ROBO2
Summary ?
| GeneID | 6092 |
| Symbol | ROBO2 |
| Synonyms | SAX3 |
| Description | roundabout guidance receptor 2 |
| Reference | MIM:602431|HGNC:HGNC:10250|Ensembl:ENSG00000185008|Vega:OTTHUMG00000158935 |
| Gene type | protein-coding |
| Map location | 3p12.3 |
| Fetal beta | 1.021 |
| eGene | Myers' cis & trans |
| Support | CELL ADHESION AND TRANSSYNAPTIC SIGNALING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.04047 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.04359 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9882137 | chr3 | 29494146 | ROBO2 | 6092 | 0.02 | trans | ||
| rs9882501 | chr3 | 29494429 | ROBO2 | 6092 | 0.16 | trans | ||
| rs4491879 | chr3 | 189373908 | ROBO2 | 6092 | 0.05 | trans | ||
| rs4505678 | chr3 | 189374356 | ROBO2 | 6092 | 0.08 | trans | ||
| rs9368571 | chr6 | 28677757 | ROBO2 | 6092 | 0.17 | trans | ||
| rs9257144 | chr6 | 28729980 | ROBO2 | 6092 | 0.11 | trans | ||
| rs16894557 | chr6 | 28999825 | ROBO2 | 6092 | 0.01 | trans | ||
| rs1503026 | chr18 | 70842605 | ROBO2 | 6092 | 0.16 | trans | ||
| rs12326929 | chr18 | 70864592 | ROBO2 | 6092 | 0.1 | trans | ||
| rs12327665 | chr19 | 39622639 | ROBO2 | 6092 | 0.07 | trans |
Section II. Transcriptome annotation
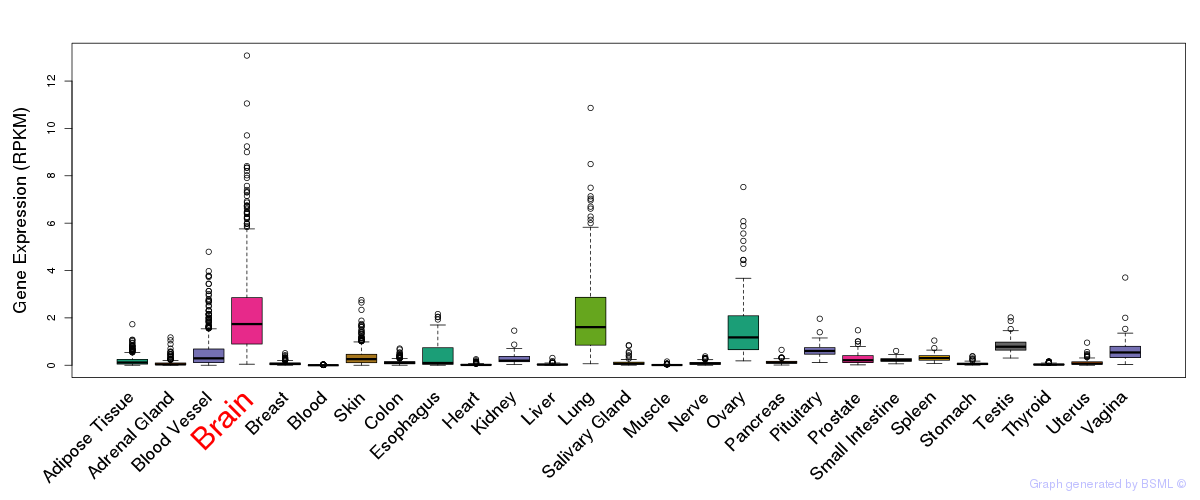
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TUT1 | 0.99 | 0.95 |
| EIF3H | 0.98 | 0.93 |
| IGBP1 | 0.97 | 0.94 |
| EIF3L | 0.97 | 0.95 |
| NACA | 0.96 | 0.95 |
| AC005393.1 | 0.96 | 0.91 |
| BTF3 | 0.96 | 0.97 |
| RPS8 | 0.96 | 0.88 |
| VPS72 | 0.96 | 0.93 |
| RPL5 | 0.96 | 0.94 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.70 | -0.76 |
| AIFM3 | -0.68 | -0.75 |
| FBXO2 | -0.68 | -0.66 |
| C5orf53 | -0.67 | -0.67 |
| LDHD | -0.66 | -0.67 |
| TINAGL1 | -0.65 | -0.77 |
| AF347015.27 | -0.65 | -0.81 |
| ALDOC | -0.65 | -0.68 |
| CLU | -0.65 | -0.68 |
| PTH1R | -0.65 | -0.69 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008046 | axon guidance receptor activity | NAS | axon (GO term level: 6) | 9458045 |
| GO:0042802 | identical protein binding | IDA | 12504588 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007420 | brain development | IEP | Brain (GO term level: 7) | 10197527 |
| GO:0050772 | positive regulation of axonogenesis | IDA | axon, neurogenesis (GO term level: 14) | 12504588 |
| GO:0001657 | ureteric bud development | IMP | 15130495 | |
| GO:0007155 | cell adhesion | IEA | - | |
| GO:0007156 | homophilic cell adhesion | IDA | 12504588 | |
| GO:0006935 | chemotaxis | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0009986 | cell surface | IDA | 12504588 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG AXON GUIDANCE | 129 | 103 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ROBO RECEPTOR | 30 | 26 | All SZGR 2.0 genes in this pathway |
| LI CISPLATIN RESISTANCE DN | 35 | 20 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY UP | 250 | 168 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER BY MUTATION RATE | 20 | 18 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF ICP WITH H3K4ME3 AND H3K27ME3 | 38 | 34 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS ICP WITH H3K4ME3 AND H327ME3 | 126 | 83 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| HOELZEL NF1 TARGETS DN | 115 | 73 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 513 | 520 | 1A,m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-130/301 | 428 | 434 | m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-144 | 514 | 520 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-148/152 | 429 | 435 | m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-153 | 296 | 303 | 1A,m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-18 | 190 | 196 | 1A | hsa-miR-18a | UAAGGUGCAUCUAGUGCAGAUA |
| hsa-miR-18b | UAAGGUGCAUCUAGUGCAGUUA | ||||
| miR-181 | 493 | 499 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-185 | 934 | 940 | 1A | hsa-miR-185brain | UGGAGAGAAAGGCAGUUC |
| miR-19 | 427 | 433 | m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-218 | 453 | 460 | 1A,m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-23 | 495 | 502 | 1A,m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC | ||||
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-25/32/92/363/367 | 76 | 82 | m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-323 | 495 | 501 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-33 | 115 | 121 | 1A | hsa-miR-33 | GUGCAUUGUAGUUGCAUUG |
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| miR-378 | 207 | 213 | m8 | hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU |
| miR-448 | 297 | 303 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-496 | 121 | 127 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.