| KEGG ERBB SIGNALING PATHWAY
| 87 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG MTOR SIGNALING PATHWAY
| 52 | 40 | All SZGR 2.0 genes in this pathway |
| KEGG TGF BETA SIGNALING PATHWAY
| 86 | 64 | All SZGR 2.0 genes in this pathway |
| KEGG FC GAMMA R MEDIATED PHAGOCYTOSIS
| 97 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY
| 137 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG ACUTE MYELOID LEUKEMIA
| 60 | 47 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAPK PATHWAY
| 87 | 68 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF
| 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY SCF KIT
| 78 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4
| 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2
| 101 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER
| 109 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB4 SIGNALING
| 38 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB2 SIGNALING
| 44 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR
| 97 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY THE B CELL RECEPTOR BCR
| 126 | 90 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE
| 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE
| 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K AKT ACTIVATION
| 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME GAB1 SIGNALOSOME
| 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF
| 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNAL TRANSDUCTION
| 95 | 76 | All SZGR 2.0 genes in this pathway |
| REACTOME PI 3K CASCADE
| 56 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR
| 100 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM
| 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM
| 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME PIP3 ACTIVATES AKT SIGNALING
| 29 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR
| 112 | 80 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN
| 309 | 191 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP
| 309 | 199 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK
| 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN
| 637 | 377 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS
| 957 | 597 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS MODERATELY DN
| 110 | 64 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 240 HELA
| 60 | 43 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 11Q12 Q14 AMPLICON
| 158 | 93 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION
| 405 | 264 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON UP
| 77 | 47 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN
| 841 | 431 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS UP
| 295 | 155 | All SZGR 2.0 genes in this pathway |
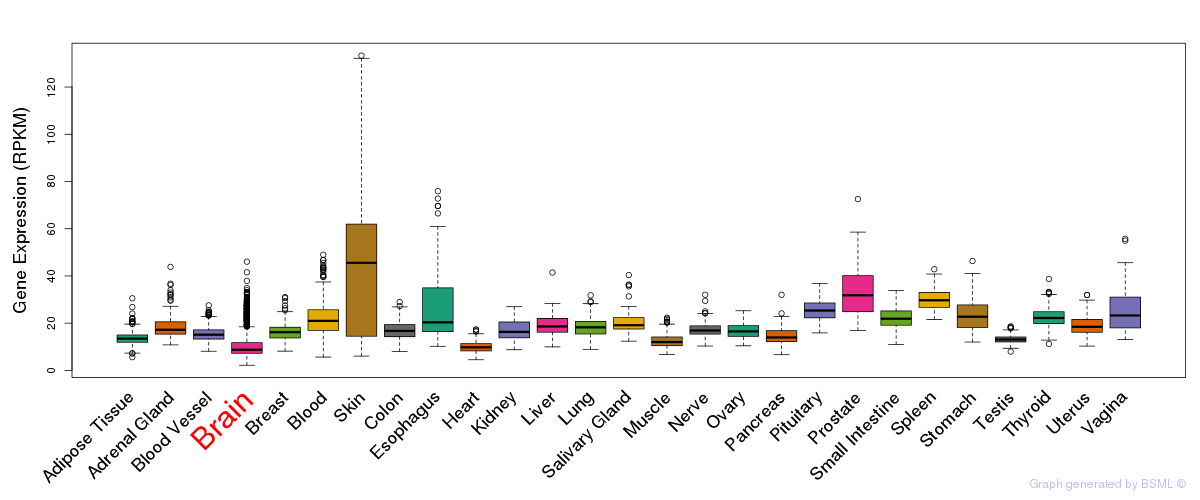
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation