Gene Page: RPS15
Summary ?
| GeneID | 6209 |
| Symbol | RPS15 |
| Synonyms | RIG|S15 |
| Description | ribosomal protein S15 |
| Reference | MIM:180535|HGNC:HGNC:10388|Ensembl:ENSG00000115268|HPRD:01613|Vega:OTTHUMG00000140099 |
| Gene type | protein-coding |
| Map location | 19p13.3 |
| Pascal p-value | 0.037 |
| Sherlock p-value | 0.957 |
| Fetal beta | 0.273 |
| DMG | 1 (# studies) |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15408407 | 19 | 1438438 | RPS15 | -0.036 | 0.41 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
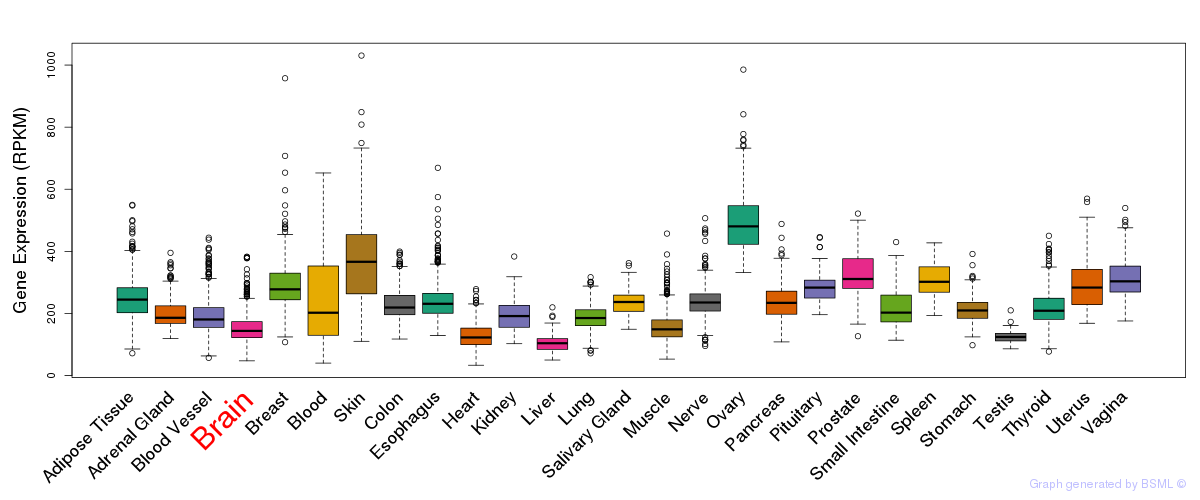
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DBNDD2 | 0.93 | 0.93 |
| APOD | 0.93 | 0.92 |
| CYP27A1 | 0.92 | 0.94 |
| MAL | 0.92 | 0.93 |
| PLLP | 0.92 | 0.92 |
| PLA2G16 | 0.91 | 0.95 |
| SEMA3B | 0.91 | 0.88 |
| SEPT4 | 0.91 | 0.89 |
| S1PR5 | 0.90 | 0.90 |
| MAG | 0.90 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NKIRAS2 | -0.63 | -0.70 |
| CRMP1 | -0.61 | -0.70 |
| KIAA1949 | -0.61 | -0.68 |
| TUBB2B | -0.60 | -0.71 |
| ZBTB8A | -0.60 | -0.69 |
| HN1 | -0.60 | -0.71 |
| ZNF821 | -0.60 | -0.69 |
| STMN1 | -0.60 | -0.71 |
| PCDHB18 | -0.60 | -0.69 |
| STMN2 | -0.60 | -0.70 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG RIBOSOME | 88 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSLATION | 222 | 75 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | 74 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | 179 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | 84 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE CHAIN ELONGATION | 153 | 41 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | 176 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLUENZA LIFE CYCLE | 203 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | 169 | 47 | All SZGR 2.0 genes in this pathway |
| REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | 176 | 57 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA DN | 116 | 79 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| TONG INTERACT WITH PTTG1 | 57 | 32 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE UP | 157 | 105 | All SZGR 2.0 genes in this pathway |
| ABE INNER EAR | 48 | 26 | All SZGR 2.0 genes in this pathway |
| CAMPS COLON CANCER COPY NUMBER UP | 92 | 45 | All SZGR 2.0 genes in this pathway |
| CHNG MULTIPLE MYELOMA HYPERPLOID UP | 52 | 25 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| BILANGES RAPAMYCIN SENSITIVE GENES | 39 | 20 | All SZGR 2.0 genes in this pathway |
| BOUDOUKHA BOUND BY IGF2BP2 | 111 | 59 | All SZGR 2.0 genes in this pathway |