Gene Page: MAPK12
Summary ?
| GeneID | 6300 |
| Symbol | MAPK12 |
| Synonyms | ERK-6|ERK3|ERK6|MAPK 12|P38GAMMA|PRKM12|SAPK-3|SAPK3 |
| Description | mitogen-activated protein kinase 12 |
| Reference | MIM:602399|HGNC:HGNC:6874|Ensembl:ENSG00000188130|HPRD:03868|Vega:OTTHUMG00000030145 |
| Gene type | protein-coding |
| Map location | 22q13.33 |
| Pascal p-value | 0.014 |
| Fetal beta | -0.321 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15134628 | 22 | 50699649 | MAPK12 | -0.023 | 0.27 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs199505423 | 22 | 50670801 | MAPK12 | ENSG00000188130.9 | 1.674E-6 | 0 | 29453 | gtex_brain_ba24 |
| rs13055135 | 22 | 50672479 | MAPK12 | ENSG00000188130.9 | 3.453E-7 | 0 | 27775 | gtex_brain_ba24 |
| rs67157296 | 22 | 50672877 | MAPK12 | ENSG00000188130.9 | 6.05E-7 | 0 | 27377 | gtex_brain_ba24 |
| rs67881086 | 22 | 50673439 | MAPK12 | ENSG00000188130.9 | 3.334E-7 | 0 | 26815 | gtex_brain_ba24 |
| rs5771256 | 22 | 50674232 | MAPK12 | ENSG00000188130.9 | 1.004E-6 | 0 | 26022 | gtex_brain_ba24 |
| rs5771258 | 22 | 50675648 | MAPK12 | ENSG00000188130.9 | 9.913E-7 | 0 | 24606 | gtex_brain_ba24 |
| rs9628271 | 22 | 50676459 | MAPK12 | ENSG00000188130.9 | 4.893E-7 | 0 | 23795 | gtex_brain_ba24 |
| rs6010213 | 22 | 50676839 | MAPK12 | ENSG00000188130.9 | 5.548E-7 | 0 | 23415 | gtex_brain_ba24 |
| rs200713867 | 22 | 50676973 | MAPK12 | ENSG00000188130.9 | 1.826E-6 | 0 | 23281 | gtex_brain_ba24 |
| rs6010216 | 22 | 50679471 | MAPK12 | ENSG00000188130.9 | 5.97E-8 | 0 | 20783 | gtex_brain_ba24 |
| rs7290465 | 22 | 50679758 | MAPK12 | ENSG00000188130.9 | 3.087E-8 | 0 | 20496 | gtex_brain_ba24 |
| rs200968480 | 22 | 50682063 | MAPK12 | ENSG00000188130.9 | 3.68E-8 | 0 | 18191 | gtex_brain_ba24 |
| rs7292864 | 22 | 50683792 | MAPK12 | ENSG00000188130.9 | 6.335E-8 | 0 | 16462 | gtex_brain_ba24 |
| rs13058062 | 22 | 50684296 | MAPK12 | ENSG00000188130.9 | 5.796E-8 | 0 | 15958 | gtex_brain_ba24 |
| rs738334 | 22 | 50688235 | MAPK12 | ENSG00000188130.9 | 6.152E-13 | 0 | 12019 | gtex_brain_ba24 |
| rs1555048 | 22 | 50688348 | MAPK12 | ENSG00000188130.9 | 5.7E-13 | 0 | 11906 | gtex_brain_ba24 |
| rs909688 | 22 | 50689030 | MAPK12 | ENSG00000188130.9 | 1.948E-13 | 0 | 11224 | gtex_brain_ba24 |
| rs2341111 | 22 | 50690322 | MAPK12 | ENSG00000188130.9 | 1.675E-13 | 0 | 9932 | gtex_brain_ba24 |
| rs5771276 | 22 | 50692562 | MAPK12 | ENSG00000188130.9 | 2.964E-12 | 0 | 7692 | gtex_brain_ba24 |
| rs2009816 | 22 | 50692848 | MAPK12 | ENSG00000188130.9 | 1.037E-12 | 0 | 7406 | gtex_brain_ba24 |
| rs2880034 | 22 | 50692853 | MAPK12 | ENSG00000188130.9 | 1.876E-8 | 0 | 7401 | gtex_brain_ba24 |
| rs2009815 | 22 | 50692856 | MAPK12 | ENSG00000188130.9 | 7.845E-10 | 0 | 7398 | gtex_brain_ba24 |
| rs4077315 | 22 | 50692908 | MAPK12 | ENSG00000188130.9 | 3.959E-9 | 0 | 7346 | gtex_brain_ba24 |
| rs2002766 | 22 | 50692966 | MAPK12 | ENSG00000188130.9 | 1.648E-11 | 0 | 7288 | gtex_brain_ba24 |
| rs5771280 | 22 | 50693340 | MAPK12 | ENSG00000188130.9 | 5.21E-14 | 0 | 6914 | gtex_brain_ba24 |
| rs5771281 | 22 | 50693479 | MAPK12 | ENSG00000188130.9 | 5.206E-14 | 0 | 6775 | gtex_brain_ba24 |
| rs5771282 | 22 | 50693521 | MAPK12 | ENSG00000188130.9 | 4.042E-12 | 0 | 6733 | gtex_brain_ba24 |
| rs6010218 | 22 | 50693534 | MAPK12 | ENSG00000188130.9 | 8.961E-12 | 0 | 6720 | gtex_brain_ba24 |
| rs6010219 | 22 | 50693538 | MAPK12 | ENSG00000188130.9 | 9.361E-11 | 0 | 6716 | gtex_brain_ba24 |
| rs6010220 | 22 | 50693554 | MAPK12 | ENSG00000188130.9 | 1.218E-13 | 0 | 6700 | gtex_brain_ba24 |
| rs1129880 | 22 | 50694297 | MAPK12 | ENSG00000188130.9 | 6.495E-12 | 0 | 5957 | gtex_brain_ba24 |
| rs5771284 | 22 | 50695270 | MAPK12 | ENSG00000188130.9 | 6.474E-12 | 0 | 4984 | gtex_brain_ba24 |
| rs5771285 | 22 | 50695908 | MAPK12 | ENSG00000188130.9 | 3.992E-13 | 0 | 4346 | gtex_brain_ba24 |
| rs5771111 | 22 | 50695946 | MAPK12 | ENSG00000188130.9 | 5.147E-14 | 0 | 4308 | gtex_brain_ba24 |
| rs5771258 | 22 | 50675648 | MAPK12 | ENSG00000188130.9 | 1.396E-6 | 0 | 24606 | gtex_brain_putamen_basal |
| rs7290465 | 22 | 50679758 | MAPK12 | ENSG00000188130.9 | 1.908E-6 | 0 | 20496 | gtex_brain_putamen_basal |
| rs7292864 | 22 | 50683792 | MAPK12 | ENSG00000188130.9 | 4.098E-7 | 0 | 16462 | gtex_brain_putamen_basal |
| rs13058062 | 22 | 50684296 | MAPK12 | ENSG00000188130.9 | 5.641E-8 | 0 | 15958 | gtex_brain_putamen_basal |
| rs738334 | 22 | 50688235 | MAPK12 | ENSG00000188130.9 | 3.187E-8 | 0 | 12019 | gtex_brain_putamen_basal |
| rs1555048 | 22 | 50688348 | MAPK12 | ENSG00000188130.9 | 3.066E-8 | 0 | 11906 | gtex_brain_putamen_basal |
| rs2341111 | 22 | 50690322 | MAPK12 | ENSG00000188130.9 | 1.578E-8 | 0 | 9932 | gtex_brain_putamen_basal |
| rs5771276 | 22 | 50692562 | MAPK12 | ENSG00000188130.9 | 1.621E-9 | 0 | 7692 | gtex_brain_putamen_basal |
| rs2009816 | 22 | 50692848 | MAPK12 | ENSG00000188130.9 | 2.11E-8 | 0 | 7406 | gtex_brain_putamen_basal |
| rs2880034 | 22 | 50692853 | MAPK12 | ENSG00000188130.9 | 1.233E-8 | 0 | 7401 | gtex_brain_putamen_basal |
| rs2009815 | 22 | 50692856 | MAPK12 | ENSG00000188130.9 | 3.79E-9 | 0 | 7398 | gtex_brain_putamen_basal |
| rs4077315 | 22 | 50692908 | MAPK12 | ENSG00000188130.9 | 2.837E-8 | 0 | 7346 | gtex_brain_putamen_basal |
| rs2002766 | 22 | 50692966 | MAPK12 | ENSG00000188130.9 | 2.04E-9 | 0 | 7288 | gtex_brain_putamen_basal |
| rs5771280 | 22 | 50693340 | MAPK12 | ENSG00000188130.9 | 4.67E-9 | 0 | 6914 | gtex_brain_putamen_basal |
| rs5771281 | 22 | 50693479 | MAPK12 | ENSG00000188130.9 | 4.655E-9 | 0 | 6775 | gtex_brain_putamen_basal |
| rs6010218 | 22 | 50693534 | MAPK12 | ENSG00000188130.9 | 1.384E-8 | 0 | 6720 | gtex_brain_putamen_basal |
| rs6010219 | 22 | 50693538 | MAPK12 | ENSG00000188130.9 | 1.219E-9 | 0 | 6716 | gtex_brain_putamen_basal |
| rs6010220 | 22 | 50693554 | MAPK12 | ENSG00000188130.9 | 5.11E-9 | 0 | 6700 | gtex_brain_putamen_basal |
| rs1129880 | 22 | 50694297 | MAPK12 | ENSG00000188130.9 | 1.225E-8 | 0 | 5957 | gtex_brain_putamen_basal |
| rs5771284 | 22 | 50695270 | MAPK12 | ENSG00000188130.9 | 9.217E-9 | 0 | 4984 | gtex_brain_putamen_basal |
| rs5771285 | 22 | 50695908 | MAPK12 | ENSG00000188130.9 | 5.183E-9 | 0 | 4346 | gtex_brain_putamen_basal |
| rs5771111 | 22 | 50695946 | MAPK12 | ENSG00000188130.9 | 4.597E-9 | 0 | 4308 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
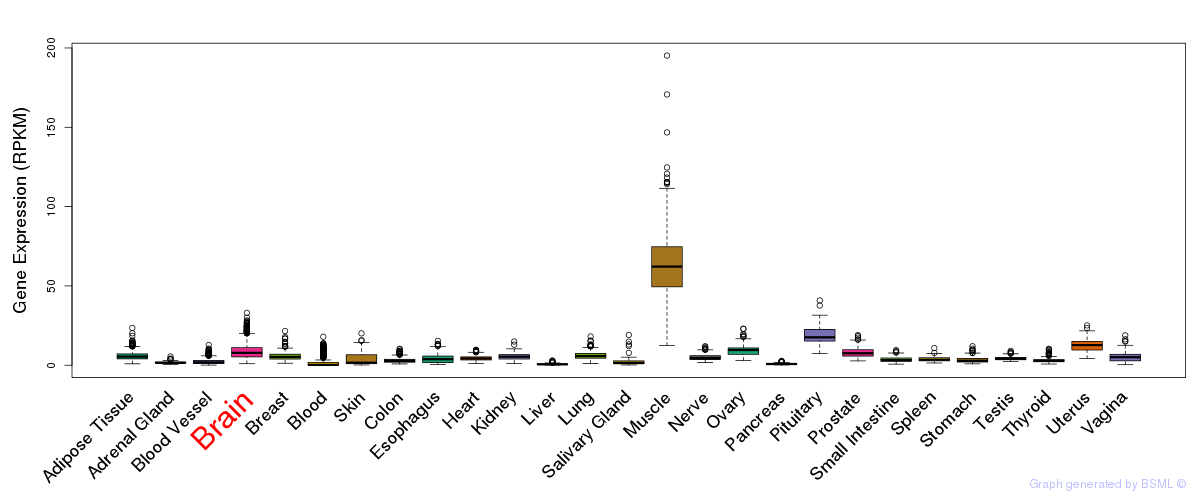
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| REV3L | 0.86 | 0.91 |
| RHOBTB1 | 0.83 | 0.84 |
| HMGCS1 | 0.82 | 0.88 |
| NFE2L3 | 0.81 | 0.88 |
| L3MBTL3 | 0.81 | 0.90 |
| SCN3A | 0.81 | 0.91 |
| KIAA0895 | 0.81 | 0.88 |
| DISP1 | 0.80 | 0.85 |
| FAM161A | 0.80 | 0.87 |
| ZBTB8B | 0.79 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.59 | -0.76 |
| REEP6 | -0.57 | -0.56 |
| AIFM3 | -0.56 | -0.72 |
| CA4 | -0.56 | -0.73 |
| TSC22D4 | -0.56 | -0.75 |
| ALDOC | -0.56 | -0.68 |
| AF347015.31 | -0.55 | -0.80 |
| FBXO2 | -0.55 | -0.61 |
| S100B | -0.55 | -0.75 |
| AF347015.27 | -0.55 | -0.78 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| KEGG VEGF SIGNALING PATHWAY | 76 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| KEGG NOD LIKE RECEPTOR SIGNALING PATHWAY | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG RIG I LIKE RECEPTOR SIGNALING PATHWAY | 71 | 51 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| KEGG FC EPSILON RI SIGNALING PATHWAY | 79 | 58 | All SZGR 2.0 genes in this pathway |
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG GNRH SIGNALING PATHWAY | 101 | 77 | All SZGR 2.0 genes in this pathway |
| KEGG PROGESTERONE MEDIATED OOCYTE MATURATION | 86 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG AMYOTROPHIC LATERAL SCLEROSIS ALS | 53 | 43 | All SZGR 2.0 genes in this pathway |
| KEGG EPITHELIAL CELL SIGNALING IN HELICOBACTER PYLORI INFECTION | 68 | 44 | All SZGR 2.0 genes in this pathway |
| KEGG LEISHMANIA INFECTION | 72 | 56 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAPK PATHWAY | 87 | 68 | All SZGR 2.0 genes in this pathway |
| SIG CD40PATHWAYMAP | 34 | 28 | All SZGR 2.0 genes in this pathway |
| ST P38 MAPK PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| PID RHOA PATHWAY | 45 | 33 | All SZGR 2.0 genes in this pathway |
| PID P38 GAMMA DELTA PATHWAY | 11 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING TO RAS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING TO ERKS | 36 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME P38MAPK EVENTS | 13 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME MYOGENESIS | 28 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME DSCAM INTERACTIONS | 11 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME NOD1 2 SIGNALING PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | 46 | 33 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F UP | 185 | 119 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| GOUYER TATI TARGETS DN | 17 | 12 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 22Q13 AMPLICON | 17 | 15 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| GOLDRATH IMMUNE MEMORY | 65 | 42 | All SZGR 2.0 genes in this pathway |
| ABRAHAM ALPC VS MULTIPLE MYELOMA UP | 26 | 22 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS UP | 113 | 71 | All SZGR 2.0 genes in this pathway |
| TSENG ADIPOGENIC POTENTIAL UP | 30 | 19 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| WOO LIVER CANCER RECURRENCE UP | 105 | 75 | All SZGR 2.0 genes in this pathway |
| KESHELAVA MULTIPLE DRUG RESISTANCE | 88 | 56 | All SZGR 2.0 genes in this pathway |