Gene Page: SDF2
Summary ?
| GeneID | 6388 |
| Symbol | SDF2 |
| Synonyms | - |
| Description | stromal cell derived factor 2 |
| Reference | MIM:602934|HGNC:HGNC:10675|Ensembl:ENSG00000132581|HPRD:04243|Vega:OTTHUMG00000132681 |
| Gene type | protein-coding |
| Map location | 17q11.2 |
| Pascal p-value | 0.189 |
| Sherlock p-value | 0.167 |
| Fetal beta | 0.085 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg22463517 | 17 | 26988906 | SDF2 | 2.55E-10 | -0.011 | 6.32E-7 | DMG:Jaffe_2016 |
| cg10824491 | 17 | 26989063 | SDF2 | 1.34E-8 | -0.006 | 5.33E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
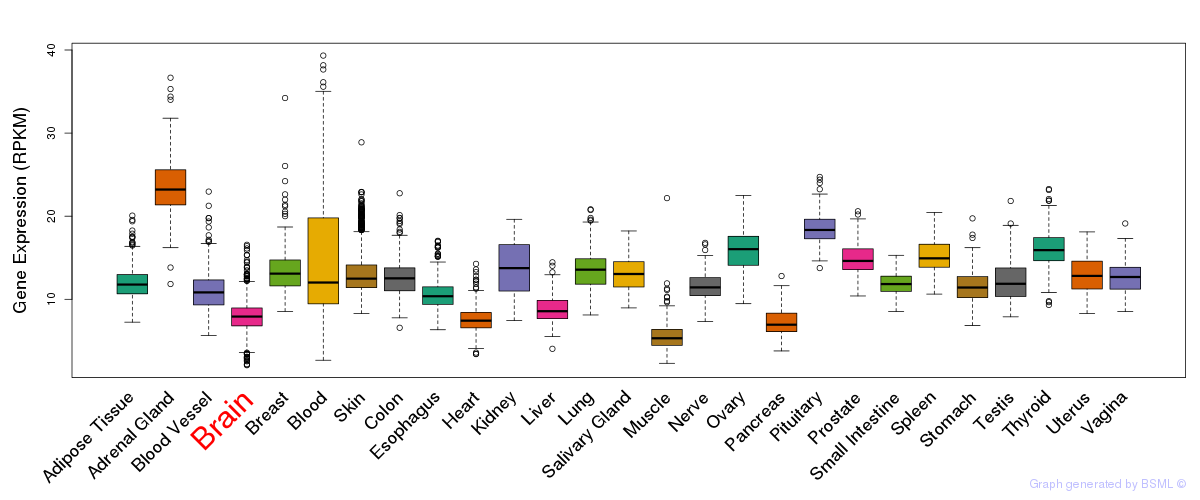
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CD44 | 0.82 | 0.73 |
| ECM2 | 0.79 | 0.73 |
| AK7 | 0.74 | 0.64 |
| ANXA1 | 0.73 | 0.63 |
| CCDC151 | 0.72 | 0.65 |
| AC079354.2 | 0.71 | 0.60 |
| DNAH9 | 0.71 | 0.67 |
| MYLK3 | 0.71 | 0.48 |
| EFHC2 | 0.70 | 0.61 |
| RSPH1 | 0.70 | 0.77 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLA | -0.40 | -0.42 |
| ISLR2 | -0.39 | -0.45 |
| DACT1 | -0.39 | -0.43 |
| KLHL1 | -0.38 | -0.44 |
| MPPED1 | -0.38 | -0.48 |
| SSBP2 | -0.38 | -0.47 |
| TMEM108 | -0.38 | -0.41 |
| SCUBE1 | -0.38 | -0.49 |
| SATB2 | -0.38 | -0.45 |
| VASH2 | -0.37 | -0.44 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 1 | 45 | 27 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP | 309 | 199 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| MORI MATURE B LYMPHOCYTE UP | 90 | 62 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q11 Q21 AMPLICON | 133 | 78 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION DN | 128 | 90 | All SZGR 2.0 genes in this pathway |
| XU CREBBP TARGETS DN | 44 | 31 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART ATRIUM DN | 141 | 99 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| MAEKAWA ATF2 TARGETS | 24 | 19 | All SZGR 2.0 genes in this pathway |
| BOUDOUKHA BOUND BY IGF2BP2 | 111 | 59 | All SZGR 2.0 genes in this pathway |