Gene Page: SGTA
Summary ?
| GeneID | 6449 |
| Symbol | SGTA |
| Synonyms | SGT|alphaSGT|hSGT |
| Description | small glutamine rich tetratricopeptide repeat containing alpha |
| Reference | MIM:603419|HGNC:HGNC:10819|Ensembl:ENSG00000104969|HPRD:04568|Vega:OTTHUMG00000180474 |
| Gene type | protein-coding |
| Map location | 19p13 |
| Pascal p-value | 1.26E-4 |
| Sherlock p-value | 0.995 |
| Fetal beta | 0.443 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg19693284 | 19 | 2783607 | SGTA | 2.01E-8 | -0.011 | 6.99E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7670908 | chr4 | 110892624 | SGTA | 6449 | 0.09 | trans | ||
| rs9991367 | chr4 | 110894300 | SGTA | 6449 | 0.09 | trans | ||
| rs1578978 | chr8 | 118196802 | SGTA | 6449 | 0.09 | trans |
Section II. Transcriptome annotation
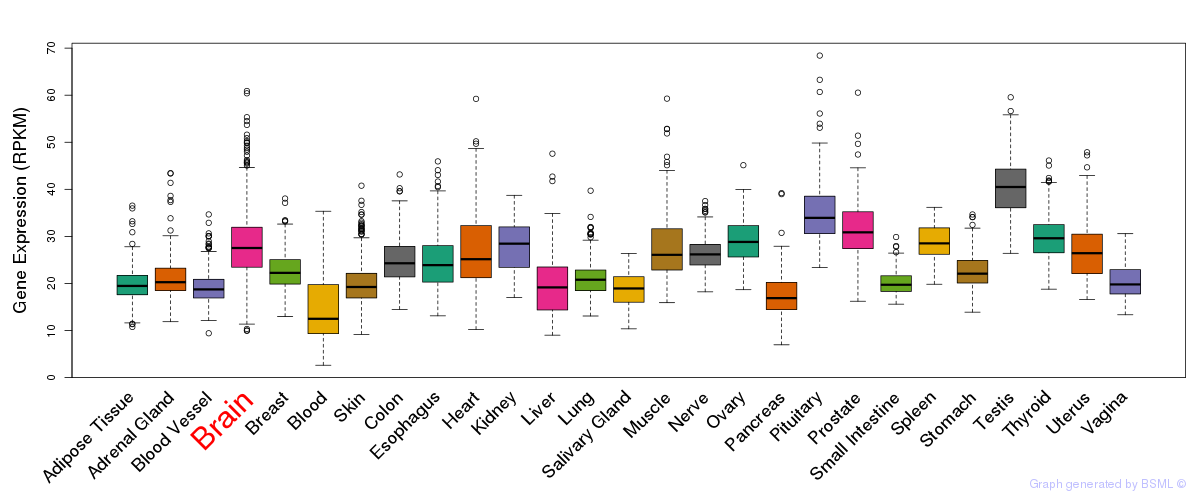
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| C1QTNF1 | CTRP1 | FLJ90694 | GIP | ZSIG37 | C1q and tumor necrosis factor related protein 1 | Two-hybrid | BioGRID | 16189514 |
| EFEMP1 | DHRD | DRAD | FBLN3 | FBNL | FLJ35535 | MGC111353 | MLVT | MTLV | S1-5 | EGF-containing fibulin-like extracellular matrix protein 1 | Two-hybrid | BioGRID | 16189514 |
| EFEMP2 | FBLN4 | MBP1 | UPH1 | EGF-containing fibulin-like extracellular matrix protein 2 | Two-hybrid | BioGRID | 16189514 |
| F11R | CD321 | JAM | JAM-1 | JAM-A | JAM1 | JAMA | JCAM | KAT | PAM-1 | F11 receptor | Two-hybrid | BioGRID | 16189514 |
| GHR | GHBP | growth hormone receptor | - | HPRD,BioGRID | 12735788 |
| GPX7 | CL683 | FLJ14777 | GPX6 | NPGPx | glutathione peroxidase 7 | Two-hybrid | BioGRID | 16189514 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | - | HPRD,BioGRID | 12482202 |
| HSPA4 | APG-2 | HS24/P52 | MGC131852 | RY | hsp70 | hsp70RY | heat shock 70kDa protein 4 | Far Western | BioGRID | 12482202 |
| IGLC1 | IGLC | immunoglobulin lambda constant 1 (Mcg marker) | Two-hybrid | BioGRID | 16189514 |
| MSTN | GDF8 | myostatin | hSGT interacts with the amino terminal region of myostatin. | BIND | 14623262 |
| NME3 | DR-nm23 | KIAA0516 | NDPK-C | NDPKC | NM23-H3 | c371H6.2 | non-metastatic cells 3, protein expressed in | Two-hybrid | BioGRID | 16189514 |
| PFN2 | D3S1319E | PFL | profilin 2 | Two-hybrid | BioGRID | 16169070 |
| PTN | HARP | HBGF8 | HBNF | NEGF1 | pleiotrophin | Two-hybrid | BioGRID | 16169070 |
| SGTA | SGT | alphaSGT | hSGT | small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha | - | HPRD | 9557704 |
| SPP1 | BNSP | BSPI | ETA-1 | MGC110940 | OPN | secreted phosphoprotein 1 | Two-hybrid | BioGRID | 16189514 |
| SPPL2A | IMP3 | PSL2 | signal peptide peptidase-like 2A | Two-hybrid | BioGRID | 16189514 |
| SRGN | FLJ12930 | MGC9289 | PPG | PRG | PRG1 | serglycin | Two-hybrid | BioGRID | 16189514 |
| SYT4 | HsT1192 | KIAA1342 | synaptotagmin IV | Two-hybrid | BioGRID | 16189514 |
| ZG16 | MGC34820 | zymogen granule protein 16 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN DN | 241 | 146 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA PROGRESSION RISK | 74 | 44 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC AND SERUM RESPONSE SYNERGY | 32 | 22 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH VS LOW DN | 32 | 21 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE UP | 157 | 105 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| KIM GASTRIC CANCER CHEMOSENSITIVITY | 103 | 64 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |